Research Article
Knockdown of ERN1 Modifies the Impact of Glutamine Deprivation on TGIF1, ZEB2, NKX3-1, PRRX1, and SLC1A5 Gene Expressions in U87 Glioblastoma Cells
- Dmytro O. Minchenko 1,2
- Dariia A. Krasnytska 1
- Olena O. Khita 1
- Yulia M. Viletska 1
- Olha V. Rudnytska 1
- Halyna E. Kozynkevych 2
- Sofiia L. Hoian 1
- Oleksandr H. Minchenko 1*
1Department of Molecular Biology, Palladin Institute of Biochemistry, National Academy of Sciences of Ukraine, Ukraine.
2Department of Pediatrics, National Bohomolets Medical University, Kyiv, Ukraine.
*Corresponding Author: Oleksandr H Minchenko, Department of Molecular Biology, Palladin Institute of Biochemistry, National Academy of Sciences of Ukraine, Ukraine.
Citation: Minchenko DO, Krasnytska DA, Khita OO, Viletska YM, Minchenko OH, et.al. (2023). Knockdown of ERN1 Modifies the Impact of Glutamine Deprivation on TGIF1, ZEB2, NKX3-1, PRRX1, and SLC1A5 Gene Expressions in U87 Glioblastoma Cells, Journal of Endocrinology and Diabetes Research, BioRes Scientia Publishers. 1(1):1-10. DOI: 10.59657/2996-3095.brs.23.004
Copyright: © 2023 Oleksandr H Minchenko, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: October 16, 2023 | Accepted: November 07, 2023 | Published: November 17, 2023
Abstract
Abstract
The aim of this investigation was to study the expression of genes encoding homeobox proteins in U87 glioblastoma cells in response to glutamine deprivation in control and ERN1 knockdown glioblastoma cells for evaluation of the significance of ERN1 signaling in the control of glioblastoma growth. It was shown that the expression level of ZEB2, TGIF1, NKX3-1, and SLC1A5 genes was up-regulated in control glioblastoma cells treated by glutamine deprivation, but ERN1 knockdown modified the sensitivity of studied genes to glutamine deprivation. Thus, treatment of glioblastoma cells without ERN1 enzymatic activity by glutamine deprivation condition led to down-regulation of the expression level of ZEB2 and more significant up-regulation of TGIF1, PRRX1, and SLC1A5 genes. These results demonstrate that ERN1 controls the sensitivity of all studied genes to glutamine deprivation and that glutamine contributes to glioblastoma cell growth through ERN1.
Keywords: ern1 knockdown; homeobox genes; slc1a5; mRNA expression; glutamine deprivation; U87 glioblastoma cells
Introduction
Malignant glioblastomas are highly aggressive tumors with very poor prognosis and glutamine as a substrate for glycolysis is important for the glioblastoma development and a more aggressive behavior (Colombo et al. 2011; Daye and Wellen, 2012; Guo et al. 2016; Zhao et al. 2017; Jiang et al. 2019; Yoo et al. 2020; Minchenko et al. 2021b). Reliance on glutamine has been considered a hallmark of metabolism in numerous cancer cells, but the requirements for glutamine in cancer are heterogeneous (Cluntun et al. 2017). Furthermore, uptake of glutamine and subsequent glutaminolysis is critical for activation of the mTORC1 pathway, which regulates cell proliferation in cancer cells (van Geldermalsen et al. 2016). It is interesting to note that glutamine deprivation as well as endoplasmic reticulum stress are very important and complementary factors for tumor growth and that ERN1/IRE1 (endoplasmic reticulum to nucleus signaling 1/inositol requiring enzyme 1) mediated stress signaling can significantly modify the effects of glutamine deprivation on gene expressions (Drogat et al. 2007; Minchenko et al. 2015; Iurlaro et al. 2017; Jiang et al. 2019; Teramoto et al. 2019). Furthermore, ERN1 knockdown also modifies the impact of glucose deprivation on the expression of numerous factors related to insulin and glucocorticoid receptors (Minchenko et al. 2013, 2020, 2021a; Riabovol et al. 2019; Tsymbal et al. 2020; Krasnytska et al. 2022). It has also been shown that ERN1/XBP1 pathway is essential for the glutamine response and protection of β cells (Hassler et al. 2015). Bioinformatic profiling identifies a glutamine-related risk signature for the malignancy of glioblastoma and the survival of patients (Zhao et al. 2017). However, the detailed molecular mechanisms of the interaction of glutamine deprivation with ERN1 mediated stress signaling pathway are complex and warrant further study. Furthermore, there are data indicating that glutamine is an important factor for cancer cells chemoresistance (Awale et al. 2006). Therefore, arctigenin inhibits numerous cancer cells growth and induces tumor cell death under glutamine deprivation possibly by suppressing the endoplasmic reticulum stress (Awale et al. 2006; Kim et al. 2010; Gu et al. 2012; He et al. 2018). This antitumor agent removes the tolerance of cancer cells to deprivation of glutamine, but mechanism of suppression of chemoresistance upon this condition is unknown yet. Moreover, deprivation of glutamine in Balb 3T3 cell culture reveals its potential for treating experimental and human cancers (Rubin et al. 2019). Interestingly, low-glucose also protected primary glial cells but not six different glioblastoma and neuroblastoma cancer cell lines against the chemotherapy (Raffaghello et al. 2008).
The homeobox proteins are an important group of transcription factors related to tumorigenesis and has been largely examined (Guca et al. 2018; Le Magnen et al. 2018; Mai et al. 2018; Wang et al. 2018; Marchand et al. 2019; Li et al. 2019; Feng et al. 2021). Recently, it was shown that transcriptional inhibitor ZEB2 (zinc finger E-box binding homeobox 2) functions as oncogene in human carcinoma (Li et al. 2019). Furthermore, transcription factor ETS1 (ETS proto-oncogene 1) is coexpressed with ZEB2 and mediates epithelial-mesenchymal transition induced by ZEB2 in human tumors (Yalim-Camci et al. 2019). It was also shown that ZEB2 regulates the activity of ETS1 by direct binding to its promoter, but migration and invasion of cancer cells are regulated by ZEB2-induced activity of ETS1. At the same time, it was shown that microRNA miR-138-5p inhibits epithelial-mesenchymal transition, proliferation and metastasis of adenocarcinoma cells through targeting transcription factor ZEB2 (Zhu et al. 2019).
TGIF1 (TGFB induced factor homeobox 1) is a member of homeobox proteins, which are highly conserved transcription regulators. In addition to its role in inhibiting 9-cis-retinoic acid-dependent RXR alpha transcription activation of the retinoic acid responsive element, the protein is an active transcriptional co-repressor of SMAD2and represses TGF-beta signaling (Guca et al. 2018). Furthermore, it has been shown that upregulation of TGIF1 by carcinogen BaP is associated with cell proliferation, cell migration, tumor invasiveness, and metastasis of lung adenocarcinoma cells (Yang et al. 2018) and that silencing of TGIF1 suppresses migration, invasion, and metastasis of human breast cancer cells (Wang et al. 2018).
Homeobox-containing transcription factor NKX3-1 (NKX3A or NK3 homeobox 1) functions as a negative regulator of epithelial cell growth in prostate tissue. It is a tumor suppressor, because this transcription factor can inhibit proliferation and invasion of PC-3 prostate cancer cells (Le Magnen et al. 2018). Furthermore, NKX3-1 is required for reprogramming of induced pluripotent stem cell, can replace OCT4 and generate fully pluripotent stem cells (Mai et al. 2018). Regulation of NKX3.1 by androgens and 17beta-estradiol in prostate cancer cells suggest that it may have important regulatory roles during prostate cancer progression. More over, NKX3-1 protein controls C-MYC (Fonseca-Alves et al. 2018). Paired related homeobox 1 (PRRX1) has also relation to tumor growth (Marchand et al. 2019; Tang et al. 2019). There are data that PRRX1 may be one of the main driving forces for the cellular phenotype plasticity and tumor dormancy of head and neck squamous cell carcinoma (Tang et al. 2019). Furthermore, uptake of glutamine and subsequent glutaminolysis is critical for activation of signaling pathways, which regulates tumor cell growth. This process is largely controlled by SLC1A5 (solute carrier family 1 member 5), because this protein is responsible for the transport of glutamine, serine and some other amino acids and involved in the control of tumor growth (van Geldermalsen et al. 2016; Wang et al. 2019).
Malignant tumors use endoplasmic reticulum stress response and its signaling pathways to adapt and to enhance tumor cells proliferation under stressful environmental conditions including low glutamine (Bravo et al. 2013; Huber et al. 2013; Almanza et al. 2019). It is well known that activation of ERN1 branch of the endoplasmic reticulum stress response is tightly linked to apoptosis and to cell death, and suppression of its function has been demonstrated to result in significant anti-proliferative effect in glioblastoma growth (Auf et al. 2010, 2013; Logue et al. 2018; Hetz et al. 2019). The aim of this study was to examine the expression of genes encoding homeobox proteins in response to glutamine deprivation condition in control U87 glioblastoma cells and ERN1 knockdown cells for evaluation of possible significance of this deprivation condition in the control of homeobox gene expressions through IRE1 mediated endoplasmic reticulum stress signaling.
Materials and Methods
Cell lines and culture conditions. The glioblastoma cell line U87 was obtained from ATCC (USA) and grown in high glutamine (4.5 g/l) Dulbecco’s modified Eagle’s minimum essential medium (Gibco, Invitrogen, Carlsbad, CA, USA) supplemented with glutamine (2 mM), 10 Percentage fetal bovine serum (Equitech-Bio, Inc., USA), penicillin (100 units/ml; Gibco) and streptomycin (0.1 mg/ml; Gibco) at 37oC in incubator with 5% CO2. In this work we used two sublines of these cells described previously (Auf et al. 2013). One subline was obtained by selection of stable transfected clones with overexpression of vector pcDNA3.1, which was used for creation of dnERN1. This untreated subline of glioblastoma cells (control glioblastoma cells) was used as control 1 in the study of the effect of glutamine deprivation on the level of gene expressions. Second subline was obtained by selection of stable transfected clones with overexpression of ERN1 dominant/negative construct (dnERN1), having suppression of both the protein kinase and endoribonuclease activities of this signaling enzyme (Auf et al. 2013). It has been shown that cells with dnERN1 have a lower proliferation rate, do not express spliced XBP1, a key transcription factor in ERN1 signaling, and have not the phosphorylated isoform of ERN1 after induction of endoplasmic reticulum stress by tunicamycin (Auf et al. 2013). Both used in this study sublines of glioblastoma cells are grown in the presence of geneticin (G418) while these cells carrying empty vector pcDNA3.1 or dnERN1 construct. Glutamine deprivation condition were created by changing the complete DMEM medium into culture plates on DMEM medium without glutamine and plates were exposed to this condition for 16h.
RNA isolation. Total RNA was extracted from glioblastoma cells using the Trizol reagent according to manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA) as described previously (Auf et al. 2013).
Reverse transcription and quantitative PCR analysis. The expression levels of homeobox mRNAs as well as ACTB mRNA were measured in control U87 glioblastoma cells and cells with a deficiency of ERN1, introduced by dnERN1, by quantitative polymerase chain reaction using SYBRGreen Mix (ABgene, Thermo Fisher Scientific, Epsom, Surrey, UK) and “QuantStudio 5 Real-Time PCR System” (Applied Biosystems, USA). Thermo Scientific Verso cDNA Synthesis Kit (Germany) was used for reverse transcription as described previously (Minchenko et al. 2019). Polymerase chain reaction was performed in triplicate. The expression of beta-actin mRNA was used as control of analyzed RNA quantity. The pair of primers specific for each studied gene was received from Sigma-Aldrich (St. Louis, MO, U.S.A.) and used for quantitative polymerase chain reaction (Table 1).
Table 1: Characteristics of the primers used for quantitative real-time polymerase chain reaction.
| Gene symbol | Gene name | Primer’s sequence | Nucleotide numbers in sequence | GenBank accession number |
| ZEB2 | zinc finger E-box binding homeobox 2 | F: 5’- actcctgtctgtctcgcaaa | 3198–3217 | NM_014795.4 |
| R: 5’- gctcgataaggtggtgcttg | 3383–3364 | |||
| TGIF1 | TGFB induced factor homeobox 1 | F: 5’- acaagttacgggagagtcgg | 208–227 | NM_003244.3 |
| R: 5’- gttgccccttctccttctct | 441–422 | |||
| SLC1A5 | solute carrier family 1 member 5 | F: 5’- tcgtggagatggaggatgtg | 1495–1514 | NM_005628.3 |
| R: 5’- ttctcctccacgcacttcat | 1729–1710 | |||
| PRRX1 | paired related homeobox 1 | F: 5’- cgtacagatcctcgtccctc | 681–700 | NM_006902.5 |
| R: 5’- tccttggccttcagtctcag | 858–839 | |||
| NKX3-1 | NK3 homeobox 1 | F: 5’- aagaacctcaagctcacgga | 524–543 | NM_006167.4 |
| R: 5’- tgtcacctgagctggcatta | 591–572 | |||
| ACTB | beta-actin | F: 5’- catccgcaaagacctgtacg | 948–967 | NM_001101.5 |
| R: 5’- cctgcttgctgatccacatc | 1165–1146 |
Quantitative PCR analysis was performed using a special computer program “Differential expression calculator” and statistical analysis using Excel program and OriginPro 7.5 software as described previously (Rudnytska et al. 2021). Comparison of two means was performed by the use of two-tailed Student’s t-test. p less than 0.05 was considered significant in all cases. The values of studied gene expressions were normalized to the expression of beta-actin mRNA and represent as percent of control (100%). All values are expressed as mean ± SEM from triplicate measurements performed in 4 independent experiments. The amplified DNA fragments were also analyzed on a 3% agarose gel and that visualized by SYBR* Safe DNA Gel Stain (Life Technologies, Carlsbad, CA, USA).
Results
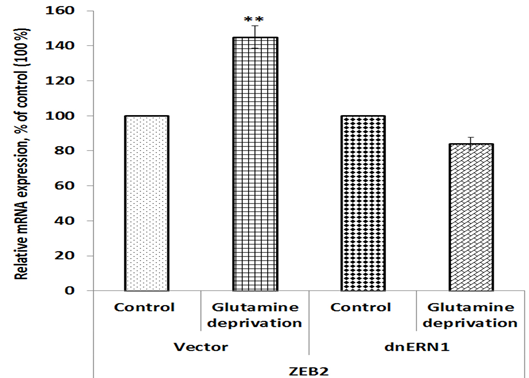
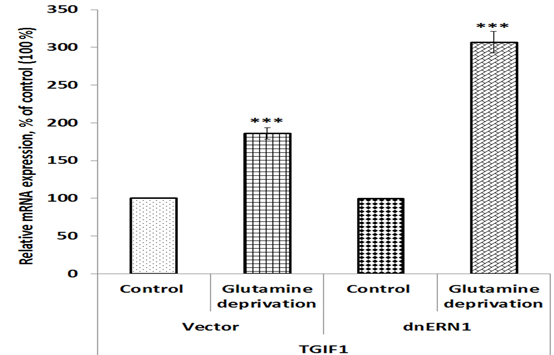
To investigate a possible role of endoplasmic reticulum stress response in the control of homeobox gene expressions in U87 glioblastoma cells under low glutamine condition we studied the impact of glutamine deprivation on the expression of homeobox genes in control glioblastoma cells (transfected by empty vector) and cells with ERN1 knockdown. As shown in Figure 1, the expression of zinc finger E-box binding homeobox 2 (ZEB2) mRNA is increased in control glioblastoma cells after exposure under condition of glutamine deprivation (+45%) in comparison with the cells growing in complete DMEM medium. Furthermore, inhibition of ERN1 signaling enzyme function by dnERN1 is significantly modified the sensitivity of ZEB2 gene expression to this experimental condition. Thus, the level of ZEB2 mRNA expression is decreased (-25%) in cells without ERN1 signaling enzyme function (Fig 1). Next, we investigated the effect of glutamine deprivation on the expression of gene encoding TGFB induced factor homeobox 1 (TGIF1) in relation to inhibition of ERN1 function. As shown in Figure 2, glutamine deprivation condition results in significant up-regulation of this homeobox gene [removed]+86%) in comparison with control glioblastoma cells, transfected by empty vector. At the same time, ERN1 knockdown leads to more strong changes in the expression of this gene (+207%; Figure 2).
Figure 1: Effect of glutamine deprivation on the expression level of zinc finger E-box binding homeobox 2 mRNA in control U87 glioblastoma cells (Vector) and cells with a blockade of the ERN1 by dnERN1 (dnERN1) measured by qPCR. Values of ZEB2 mRNA expression were normalized to beta-actin mRNA level and represented as percent for control 1 (100%); n=4.
Figure 2: Effect of glutamine deprivation on the expression level of TGFB induced factor homeobox 1 mRNA in control U87 glioblastoma cells (Vector) and cells with a blockade of the ERN1 (dnERN1) measured by qPCR. Values of TGIF1 mRNA expression were normalized to beta-actin mRNA level and represented as percent for control 1 (100%); n=4.
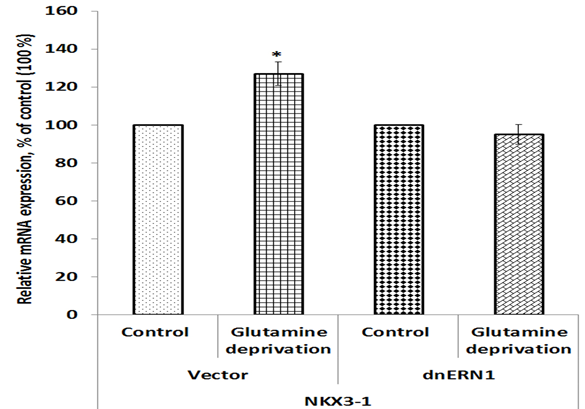
Figure 3: Effect of glutamine deprivation on the expression level of NK3 homeobox 1 mRNA in control U87 glioblastoma cells (Vector) and cells with a blockade of the ERN1 (dnERN1) measured by qPCR. Values of NKX3-1 mRNA expression were normalized to beta-actin mRNA level and represented as percent for control 1 (100%); n=4.
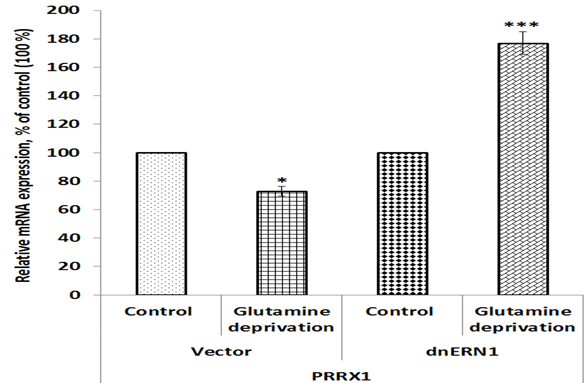
Figure 4: Effect of glutamine deprivation on the expression level of paired related homeobox 1 mRNA in control U87 glioblastoma cells (Vector) and cells with a blockade of the ERN1 (dnERN1) measured by qPCR. Values of PRRX1 mRNA expression were normalized to beta-actin mRNA level and represented as percent for control 1 (100%); n=4.
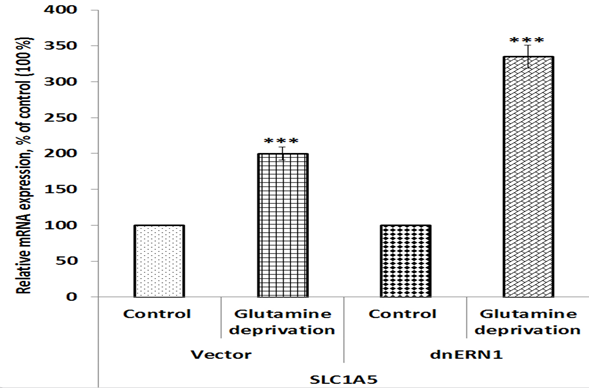
Figure 5: Effect of glutamine deprivation on the expression level of solute carrier family 1 member 5 mRNA in control U87 glioblastoma cells (Vector) and cells with a blockade of the ERN1 (dnERN1) measured by qPCR. Values of SLC1A5 mRNA expression were normalized to beta-actin mRNA level and represented as percent for control 1 (100%); NS – no significant changes; n=4.
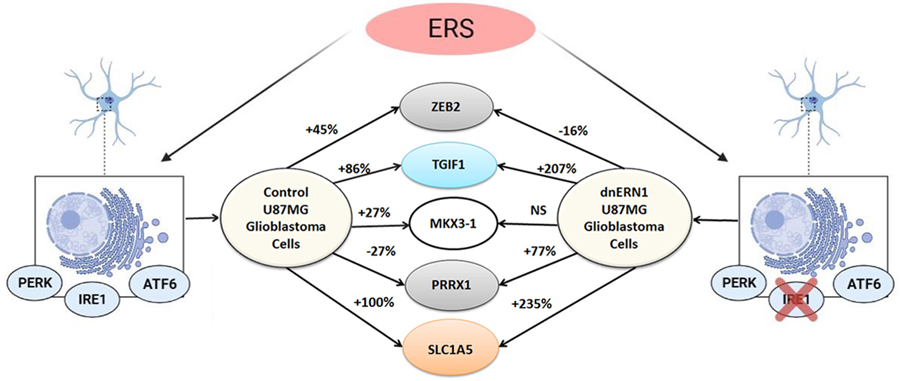
Figure 6: Schematic demonstration of changes in the expression profile of homeobox family genes in the control and ERN1 knockdown glioblastoma cells under glutamine deprivation; NS – no significant changes.
We also investigated the effect of glutamine deprivation condition on the expression of genes encoding NK3 homeobox 1 (NKX3-1) and paired related homeobox 1 (PRRX1) protein in glioblastoma cells in relation to ERN1 knockdown. As shown in Figure 3, the expression of NKX3-1 mRNA is also up-regulated in control glioblastoma cells treated by glutamine deprivation (+27%), but inhibition of ERN1 signaling protein eliminates the sensitivity of this mRNA expression to glutamine deprivation in comparison with dnERN1 cells growing with glutamine. At the same time, the expression level of PRRX1 gene is down-regulated (-27%) in control glioblastoma cells under glutamine deprivation condition in comparison with cells growing in regular medium (Figure 4). Furthermore, glutamine deprivation of ERN1 knockdown glioblastoma cells introduces the changes in this homeobox gene expression in the opposite direction (+77%) as compared to ERN1 knockdown cells growing with glutamine (Figure 4).
We have also studied the effect of glutamine deprivation on the expression of gene SLC1A5 encoding protein responsible for the transport of glutamine, serine and some other amino acids in control and ERN1 knockdown glioblastoma cells. As shown in Figure 5, exposure of glioblastoma cells under glutamine deprivation condition leads to significant up-regulation of SLC1A5 mRNA expression (+100%) in comparison with control cells growing under condition with glutamine. Furthermore, inhibition of ERN1 strongly enhances the effect of glutamine deprivation on the expression of SLC1A5 mRNA (+225%). Thus, glutamine deprivation condition affects the expression of different homeobox genes in gene-specific manner and the impact of glutamine deprivation condition on gene expressions depends on ERN1 signaling. Results of this investigation are summarized in Figure 6, which clearly demonstrated the ERN1 dependent character of changes in the expression profile of most homeobox family genes in glioblastoma cells under glutamine deprivation.
Discussion
In this work, we studied the effect of glutamine deprivation on the expression of genes encoding important homeobox proteins, which function as transcription factors, in U87 glioblastoma cells in relation to inhibition of ERN1, the major signaling pathway of the unfolded protein response. For this aim we used control glioblastoma cells, transfected by empty vector pcDNA3.1 and cells with full ERN1 deficiency introduced by dnERN1. This is important for the evaluation of possible significance of in ERN1 dependent control of glioblastoma cell proliferation because endoplasmic reticulum stress signaling mediated by ERN1 is involved in numerous metabolic pathways and ERN1 knockdown has clear anti-tumor effects (Auf et al. 2010, 2013; Bravo et al. 2013; Logue et al. 2018; Almanza et al. 2019; Minchenko et al. 2021b). Furthermore, there are data that glutamine deprivation can enhance the sensitivity of cancer cells to anti-cancer drugs, particularly arctigenin, which inhibits the growth of various cancer cells and induces tumor cell death under glutamine deprivation condition possibly by blocking the unfolded protein response and inhibiting cellular energy metabolism (Awale et al. 2006; Kim et al. 2010; Gu et al. 2012; He et al. 2018). Results of our study clarify possible mechanisms of glutamine deprivation on the proliferation/surviving of ERN1 knockdown glioblastoma cells through specific changes in the expression of genes encoding important homeobox proteins.
We showed that ERN1 knockdown of glioblastoma cells leads to a strong down-regulation of the expression of ZEB2 gene under glutamine deprivation. Homeobox protein ZEB2 is functioning as an oncogene in human laryngeal squamous cell carcinoma and acts as an upstream positive regulator of ETS1 (Li et al. 2019; Yalim-Camci et al. 2019). Furthermore, transcription factor ETS1 is coexpressed with ZEB2 and mediates ZEB2-induced epithelial-mesenchymal transition in cancers (Yalim-Camci et al. 2019). At the same time, transcription factor ZEB2 can directly bind to ETS1 promoter and control its transcription. Therefore, down-regulation of ZEB2 gene expression under glutamine deprivation may contribute to decreased proliferation potential and chemoresistance of these cells (Auf et al. 2010; Kim et al. 2010; Gu et al. 2012; Minchenko et al. 2021b).
Furthermore, we showed that the expression of TGFB induced factor homeobox 1 gene in glioblastoma cells with inhibited ERN1 signaling is strongly up-regulated under glutamine deprivation, while in control glioblastoma cells the effect of glutamine deprivation was significantly lower (Figure 6). The TGIF1 is a member of the atypical homeobox proteins, which are highly conserved transcription regulators. It is an active transcriptional co-repressor of SMAD2 and represses TGF-β signaling (Guca et al. 2018). At the same time, TGF-β signaling can suppresses tumor formation by inhibiting cell growth and apoptosis and can also promote cancer growth. It is possible that more significant induction of TGIF1 gene expression in glioblastoma cells with inhibited ERN1 signaling under glutamine deprivation condition is also contributed to suppression of glioblastoma cells proliferation by ERN1 knockdown. The NKX3-1 gene functions as a tumor suppressor and increase of its expression in control glioblastoma cells under glutamine deprivation condition possibly contribute to decreased proliferation potential of these cells (Le Magnen et al. 2018; Jiang et al. 2019). At the same time, we showed that ERN1 knockdown eliminates the sensitivity of NKX3-1 gene expressions to glutamine deprivation conditions in glioblastoma cells. It is possible that the expression of NKX3-1 is mediated by ERN1 and knockdown of ERN1 eliminates regulation of NKX3-1 expression under glutamine deprivation conditions. Furthermore, we have shown that the expression of PRRX1 gene is strongly increased in ERN1 knockdown glioblastoma cells treated by glutamine deprivation. These results agree well with suppressed proliferation of ERN1 knockdown glioblastoma cells and functions of PRRX1 as transcription repressor (Tang et al. 2019). The study of the expression of SLC1A5, which controls glutamine transport, is of great importance, as it showed not only an increase in the expression of this gene under glutamine deprivation, but also its pronounced dependence on the inhibition of ERN1. It is possible that ERN1 controls glutamine uptake and metabolism through modulation of numerous gene expressions (Hassler et al. 2015; van Geldermalsen et al. 2016; Jiang et al. 2019; Wang et al. 2019).
This study provides unique insights into the molecular mechanisms regulating the expression of genes encoding homeobox proteins in glioblastoma cells in response to glutamine deprivation and their correlation with inhibition of ERN1 activity and reduced cell proliferation in cells harboring dnERN1, attesting to the fact that endoplasmic reticulum stress as well as glutamine is a necessary component of malignant tumor growth and cell survival. Furthermore, our results validate tight interaction of endoplasmic reticulum stress signaling pathway ERN1 with glutamine deprivation in the regulation of the expression of genes encoding homeobox proteins, but the detailed molecular mechanisms of this regulation have not been yet clearly defined and requires further investigation.
Declarations
Acknowledgement
This work was funded by the State Budget Program "Support for the Development of Priority Areas of Scientific Research" (Code: 6541030).
Competing interest
All authors declare that they reviewed the manuscript and have no competing interest.
References
- Almanza A, Carlesso A, Chintha C, Creedican S, Doultsinos D, Leuzzi B, Luís A, McCarthy N, Montibeller L, More S, Papaioannou A, Püschel F, Sassano ML, Skoko J, Agostinis P, de Belleroche J, Eriksson LA, Fulda S, Gorman AM, Healy S, Kozlov A, Muñoz-Pinedo C, Rehm M, Chevet E, Samali A. (2019). Endoplasmic reticulum stress signalling - from basic mechanisms to clinical applications. FEBS J, 286;241-278.
Publisher | Google Scholor - Auf G, Jabouille A, Delugin M, Guérit S, Pineau R, North S, Platonova N, Maitre M, Favereaux A, Vajkoczy P, Seno M, Bikfalvi A, Minchenko D, Minchenko O, Moenner M. (2013). High epiregulin expression in human U87 glioma cells relies on IRE1alpha and promotes autocrine growth through EGF receptor. BMC Cancer, 13;597.
Publisher | Google Scholor - Auf G, Jabouille A, Guérit S, Pineau R, Delugin M, Bouchecareilh M, Favereaux A, Maitre M, Gaiser T, von Deimling A, Czabanka M, Vajkoczy P, Chevet E, Bikfalvi A, Moenner M. (2010). A shift from an angiogenic to invasive phenotype induced in malignant glioma by inhibition of the unfolded protein response sensor IRE1. Proc Natl Acad Sci U S A, 107;15553-15558.
Publisher | Google Scholor - Awale S, Lu J, Kalauni SK, Kurashima Y, Tezuka Y, Kadota S, Esumi H. (2006). Identification of arctigenin as an antitumor agent having the ability to eliminate the tolerance of cancer cells to nutrient starvation. Cancer Res, 66;1751-1757.
Publisher | Google Scholor - Bravo R1, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AF, Lavandero S. (2013). Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int Rev Cell Mol Biol, 301;215-290.
Publisher | Google Scholor - Cluntun AA, Lukey MJ, Cerione RA, Locasale JW. (2017). Glutamine Metabolism in Cancer: Understanding the Heterogeneity. Trends Cancer, 3;169-180.
Publisher | Google Scholor - Colombo SL, Palacios-Callender M, Frakich N, Carcamo S, Kovacs I, Tudzarova S, Moncada S. (2011). Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc Natl Acad Sci USA, 108;21069-21074.
Publisher | Google Scholor - Daye D, Wellen KE. (2012). Metabolic reprogramming in cancer: unraveling the role of glutamine in tumorigenesis. Semin Cell Dev Biol, 23;362-369.
Publisher | Google Scholor - Drogat B, Bouchecareilh M, North S, Petibois C, Déléris G, Chevet E, Bikfalvi A, Moenner M. (2007). Acute L-glutamine deprivation compromises VEGF-a upregulation in A549/8 human carcinoma cells. J Cell Physiol, 212;463-472.
Publisher | Google Scholor - Feng Y, Zhang T, Wang Y, Xie M, Ji X, Luo X, Huang W. (2021). Homeobox Genes in Cancers: From Carcinogenesis to Recent Therapeutic Intervention. Front Oncol, 11;770428.
Publisher | Google Scholor - Fonseca-Alves CE, Kobayashi PE, Laufer-Amorim R. (2018). Evaluation of NKX3.1 and C-MYC expression in canine prostatic cancer. Res Vet Sci, 118;365-370.
Publisher | Google Scholor - Gu Y, Qi C, Sun X, Ma X, Zhang H, Hu L, Yuan J, Yu Q. (2012). Arctigenin Preferentially Induces Tumor Cell Death Under Glucose Deprivation by Inhibiting Cellular Energy Metabolism. Biochem Pharmacol, 84;468-476.
Publisher | Google Scholor - Guca E, Sunol D, Ruiz L, Konkol A, Cordero J, Torner C, Aragon E, Martin-Malpartida P, Riera A, Macias MJ. (2018). TGIF1 homeodomain interacts with Smad MH1 domain and represses TGF-beta signaling. Nucleic Acids Res, 46;9220-9235.
Publisher | Google Scholor - Guo H, Nan Y, Zhen Y, Zhang Y, Guo L, Yu K, Huang Q, Zhong Y. (2016). miRNA-451 inhibits glioma cell proliferation and invasion by downregulating glutamine transporter 1. Tumour Biol, 37;13751-13761.
Publisher | Google Scholor - Hassler JR, Scheuner DL, Wang S, Han J, Kodali VK, Li P, Nguyen J, George JS, Davis C, Wu SP, Bai Y, Sartor M, Cavalcoli J, Malhi H, Baudouin G, Zhang Y, Yates Iii JR, Itkin-Ansari P, Volkmann N, Kaufman RJ. (2015). The IRE1α/XBP1s pathway is essential for the glutamine response and protection of β cells. PLoS Biol, 13;1002277.
Publisher | Google Scholor - He Y, Fan Q, Cai T, Huang W, Xie X, Wen Y, Shi Z. (2018). Molecular Mechanisms of the Action of Arctigenin in Cancer. Biomed Pharmacother, 108;403-407.
Publisher | Google Scholor - Hetz C, Axten JM, Patterson JB. (2019). Pharmacological targeting of the unfolded protein response for disease intervention. Nat Chem Biol, 15;764-775.
Publisher | Google Scholor - Huber AL, Lebeau J, Guillaumot P, Petrilli V, Malek M, Chilloux J, Fauvet F, Payen L, Kfoury A, Renno T, Chevet E, Manie SN. (2013). p58(IPK)-mediated attenuation of the proapoptotic PERK-CHOP pathway allows malignant progression under low glutamine. Mol Cell, 49;1049-1059.
Publisher | Google Scholor - Iurlaro R, Puschel F, Leon-Annicchiarico CL, O'Connor H, Martin SJ, Palou-Gramon D, Lucendo E, Munoz-Pinedo C. (2017). Glutamine deprivation induces ATF4-mediated apoptosis through TRAIL death receptors. Mol Cell Biol, 37.
Publisher | Google Scholor - Jiang J, Srivastava S, Zhang J. (2019). Starve Cancer Cells of Glutamine: Break the Spell or Make a Hungry Monster?. Cancers (Basel), 11;804.
Publisher | Google Scholor - Kim JY, Hwang JH, Cha MR, Yoon MY, Son ES, Tomida A, Ko B, Song SW, Shinya K, Hwang YI, Park HR. (2010). Arctigenin Blocks the Unfolded Protein Response and Shows Therapeutic Antitumor Activity. J Cell Physiol, 224;33-40.
Publisher | Google Scholor - Krasnytska DA, Khita OO, Tsymbal DO, Luzina OY, Cherednychenko AA, Kozynkevych HE, Bezrodny BH, Minchenko DO. (2022). The impact of glutamine deprivation on the expression of MEIS3, SPAG4, LHX1, LHX2, and LHX6 genes in ERN1 knockdown U87 glioma cells. Endocr Reg, 56;38-47.
Publisher | Google Scholor - Le Magnen C, Virk RK, Dutta A, Kim JY, Panja S, Lopez-Bujanda ZA, Califano A, Drake CG, Mitrofanova A, Abate-Shen C. (2018). Cooperation of loss of NKX3.1 and inflammation in prostate cancer initiation. Dis Model Mech, 11;35139.
Publisher | Google Scholor - Li Q, Ma L, Wu Z, Wang G, Huang Q, Shen Z, Yu R. (2019). Zinc finger Ebox binding homeobox 2 functions as an oncogene in human laryngeal squamous cell carcinoma. Mol Med Rep, 19;4545-4552.
Publisher | Google Scholor - Logue SE, McGrath EP, Cleary P, Greene S, Mnich K, Almanza A, Chevet E, Dwyer RM, Oommen A, Legembre P, Godey F, Madden EC, Leuzzi B, Obacz J, Zeng Q, Patterson JB, Jäger R, Gorman AM, Samali A. (2018). Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat Commun, 9 ;3267.
Publisher | Google Scholor - Mai T, Markov GJ, Brady JJ, Palla A, Zeng H, Sebastiano V, Blau HM. (2018). NKX3-1 is required for induced pluripotent stem cell reprogramming and can replace OCT4 in mouse and human iPSC induction. Nat Cell Biol, 20;900-908.
Publisher | Google Scholor - Marchand B, Pitarresi JR, Reichert M, Suzuki K, Laczko D, Rustgi AK. (2019). PRRX1 isoforms cooperate with FOXM1 to regulate the DNA damage response in pancreatic cancer cells. Oncogene, 38;4325-4339.
Publisher | Google Scholor - Minchenko DO, Kharkova AP, Hubenia OV, Minchenko OH. (2013). Insulin receptor, IRS1, IRS2, INSIG1, INSIG2, RRAD, and BAIAP2 gene expressions in glioma U87 cells with ERN1 loss of function: effect of hypoxia and glutamine or glucose deprivation. Endocr Reg, 47;15-26.
Publisher | Google Scholor - Minchenko DO, Kharkova AP, Tsymbal DO, Karbovskyi LL, Minchenko OH. (2015). IRE1 inhibition affects the expression of insulin-like growth factor binding protein genes and modifies its sensitivity to glucose deprivation in U87 glioma cells. Endocr Reg, 49;185-197.
Publisher | Google Scholor - Minchenko DO, Khita OO, Tsymbal DO, Danilovskyi SV, Rudnytska OV, Halkin OV, Kryvdiuk IV, Smeshkova MV, Yakymchuk MM, Bezrodnyi BH, Minchenko OH. (2020). Expression of IDE and PITRM1 genes in IRE1 knockdown U87 glioma cells: effect of hypoxia and glucose deprivation. Endocr Reg, 54;183-195.
Publisher | Google Scholor - Minchenko DO, Khita OO, Tsymbal DO, Viletska YM, Sliusar MY, Yefimova YV, Levadna LO, Krasnytska DA, Minchenko OH. (2021). ERN1 knockdown modifies the impact of glucose and glutamine deprivations on the expression of EDN1 and its receptors in glioma cells. Endocr Reg, 55;72-82.
Publisher | Google Scholor - Minchenko O.H, Tsymbal D.O, Khita O.O., Minchenko D.O. (2021). Inhibition of ERN1 signaling is important for the suppression of tumor growth. Clin Cancer Drugs, 8;27-38.
Publisher | Google Scholor - Raffaghello L, Lee C, Safdie FM, Wei M, Madia F, Bianchi G, Longo VD. (2008) Starvation-dependent differential stress resistance protects normal but not cancer cells against high-dose chemotherapy. Proc Natl Acad Sci U S A, 105;8215-8220.
Publisher | Google Scholor - Riabovol OO, Tsymbal DO, Minchenko DO, Lebid-Biletska KM, Sliusar MY, Rudnytska OV, Minchenko OH. (2019). Effect of glucose deprivation on the expression of genes encoding glucocorticoid receptor and some related factors in ERN1-knockdown U87 glioma cells. Endocr Regul, 53;237-249.
Publisher | Google Scholor - Rubin H. (2019). Deprivation of glutamine in cell culture reveals its potential for treating cancer. Proc Natl Acad Sci U S A, 116;6964-6968.
Publisher | Google Scholor - Rudnytska OV, Khita OO, Minchenko DO, Tsymbal DO, Yefimova YV, Sliusar MY, Minchenko OH. (2021). The low doses of SWCNTs exhibit a genotoxic effect on the normal human astrocytes by disrupting the functional integrity of the genome. Curr Res Toxicol, 2;64-71.
Publisher | Google Scholor - Tang Y, Lu Y, Cheng Y, Luo L, Cai L, Peng B, Huang W, Liao H, Zhao L, Pan M. (2019). Pre-metastatic niche triggers SDF-1/CXCR4 axis and promotes organ colonisation by hepatocellular circulating tumour cells via downregulation of Prrx1. J Exp Clin Cancer Res, 38;473.
Publisher | Google Scholor - Teramoto K, Katoh H. (2019). The cystine/glutamate antiporter xCT is a key regulator of EphA2 S897 phosphorylation under glutamine-limited conditions. Cell Signal, 62;109329.
Publisher | Google Scholor - Tsymbal DO, Minchenko DO, Khita OO, Rudnytska OV, Viletska YM, Lahanovska YO, He Q, Liu K, Minchenko OH. (2020). ERN1 knockdown modifies the effect of glucose deprivation on homeobox gene expressions in U87 glioma cells. Endocr Reg, 54;196-206.
Publisher | Google Scholor - van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O'Toole SA, Rasko JE, Holst J. (2016). ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene, 35;3201-3208.
Publisher | Google Scholor - Wang VM, Ferreira RMM, Almagro J, Evan T, Legrave N, Zaw Thin M, Frith D, Carvalho J, Barry DJ, Snijders AP, Herbert E, Nye EL, MacRae JI, Behrens A. (2019). CD9 identifies pancreatic cancer stem cells and modulates glutamine metabolism to fuel tumour growth. Nat Cell Biol, 21;1425-1435.
Publisher | Google Scholor - Wang Y, Li L, Wang H, Li J, Yang H. (2018). Silencing TGIF suppresses migration, invasion and metastasis of MDAMB231 human breast cancer cells. Oncol Rep, 39;802-808.
Publisher | Google Scholor - Yalim-Camci I, Balcik-Ercin P, Cetin M, Odabas G, Tokay N, Sayan AE, Yagci T. (2019). ETS1 is coexpressed with ZEB2 and mediates ZEB2-induced epithelial-mesenchymal transition in human tumors. Mol Carcinog, 58;1068-1081.
Publisher | Google Scholor - Yang H, Zhang H, Pan T, Wang H, Wang Y. (2018). Benzo(a)pyrene promotes migration, invasion and metastasis of lung adenocarcinoma cells by upregulating TGIF. Toxicol Lett, 294;11-19.
Publisher | Google Scholor - Yoo HC, Yu YC, Sung Y, Han JM. (2020). Glutamine reliance in cell metabolism. Exp Mol Med, 52;1496-1516.
Publisher | Google Scholor - Zhao S, Cai J, Li J, Bao G, Li D, Li Y, Zhai X, Jiang C, Fan L. (2017). Bioinformatic profiling identifies a glutamine-related risk signature for the malignancy of glioma and the survival of patients. Mol Neurobiol, 54;8203-8210.
Publisher | Google Scholor - Zhu D, Gu L, Li Z, Jin W, Lu Q, Ren T. (2019). MiR-138-5p suppresses lung adenocarcinoma cell epithelial-mesenchymal transition, proliferation and metastasis by targeting ZEB2. Pathol. Res Pract, 215;861-872.
Publisher | Google Scholor