Research Article
Mycobacterium Leprae Molecular Detection in Stained Slit Skin Smears from Leprosy Patients and Comparison of Molecular, Histopathological, With Clinical Data
1Hawassa University, Awasa, Ethiopia.
2Addis Ababa University, Addis Ababa, Ethiopia.
3Armauer Hansen Research Institute, Addis Ababa, Ethiopia.
4ABiTi Consultancy Services (ACS), Addis Ababa, Ethiopia.
*Corresponding Author: Giorgis Yeabyo, ABiTi Consultancy Services (ACS), Addis Ababa, Ethiopia.
Citation: Bereket A, Gemechu T, Bobosha K, Girma S, Yeabyo G. (2024). Mycobacterium Leprae Molecular Detection in Stained Slit Skin Smears from Leprosy Patients and Comparison of Molecular, Histopathological, With Clinical Data, International Journal of Clinical and Surgical Pathology, BioRes Scientia Publishers. 1(1):1-3. DOI: 10.59657/3067-0462.brs.24.001
Copyright: © 2024 Giorgis Yeabyo, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: February 20, 2024 | Accepted: June 24, 2024 | Published: July 02, 2024
Abstract
Background: Mycobacterium leprae is a causative agent of leprosy which is a chronic infectious disease. The disease mainly involves the skin and peripheral nerves. Globally the prevalence of leprosy at the end of 2015 was 210,758 cases (3.2 cases per 100, 000 populations).
Objective: To evaluate the diagnostic performance of PCR to detect M. leprae on stained Slit Skin Smears (SSS) cross-sectional collected from clinically confirmed leprosy patients.
Materials and Methods: Retrospective cross-sectional study was conducted on 60 clinically confirmed leprosy patients of 42 Multibacillary (MB) and 18 Paucibacillary (PB) leprosy cases from archival samples.
Results: The PCR on Slit Skin Smear (SSS) was positive in 6(10.00%) in PB patients and 23(38.67%) in MB patients. Among the MB and PB cases detected by PCR 3, and 2 cases respectively were from AFB negative slides.
Conclusion: The PCR on SSS detected a total of 29 out of 60 samples indicating its potential for diagnosis. Although PCR on SSS showed low detection as compared to H&E and Fite-Faraco (FF) staining (staining on punch biopsy samples), it detected more positive samples than ZN staining. Therefore, by improving some technical procedures in sample collection and handling, it can be used for diagnostics where PCR machines are available.
Keywords: slit skin smear; pb patients; schwann cells; nerves
Introduction
Mycobacterium leprae (M. leprae) is the causative agent of leprosy or Hansen’s disease which is a chronic but curable human disease affecting the skin, peripheral nerves, eyes, and mucosal surfaces of the upper respiratory tract [1]. Leprosy affects all age groups and both sexes, with the most affected being the 15–45 years age-group. M. leprae primarily invades Schwann cells in the peripheral nerves leading to nerve damage and the development of disabilities, the incubation period of the disease ranges from three to ten years [2]. M. leprae remains a diagnostic challenge in developing countries including Ethiopia for many clinicians because of the low sensitivity of conventional methods in detecting M. leprae bacilli in clinical specimens of SSS and biopsy. At present, the diagnosis of leprosy is based on the 3 cardinal signs of the disease, which are Hypo pigmented or reddish patches with definite loss of sensation; enlarged peripheral nerve, and AFB-positive slit-skin smear or biopsy [3]. The clinical and microscopic examinations are not enough for the detection of M. leprae because both lack the power to detect PB patients. The inability of the bacteria to grow in artificial media and its extremely slow generation time of 12–14 days in selected live animals are the major challenges. Therefore, there is a need for a more sensitive and faster diagnostic tool for early diagnosis of leprosy cases to prevent deformities and disabilities [4].
Materials and Methods
The samples were previously collected from ALERT hospital from new untreated patients. SSS was collected from three different body parts of the leprosy patients to increase the probability of detecting acid-fast bacilli. ZN staining was performed and done and slides were stored at the Armauer Hansen Research Institute (AHRI) Histopathology laboratory. One hundred thirty-seven clinically diagnosed PB and MB leprosy cases of archival SSS ZN stained slides were examined and stored from 2015 to 2016. Out of 137, 60 were selected from archival SSS ZN stained slides for this study. The sampling technique was purposive sampling where all forms of leprosy were represented and a retrospective cross-sectional study was conducted from February to May 2017. All data including the clinical data and Histopathological data of the PB and MB leprosy cases of 60 archived samples from 2015 to 2016 were collected and organized using the structured checklist in AHRI Histopathology laboratory. Specimen processing: DNA has been extracted from ZN-stained slides of SSS with DNeasy Blood and Tissue kit (QIAGEN, Valencia, CA, USA (Azevedo MC, et al, 2016). The oil was removed by xylene from ZN-stained slide and air dry completely, then added molecular grade water 100µl to facilitate sample release and scrap the smear by a new surgical blade. The scrapped sample was transferred to a new Eppendorf (1.5 ml microcentrifuge tube) and then processed as the Standard Operating Procedure (SOP).
Result
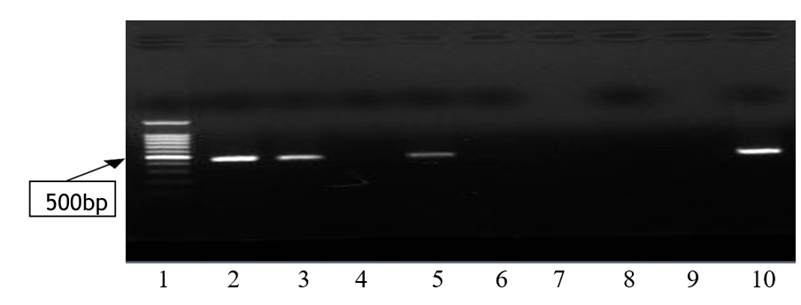
Out of the 60 clinically diagnosed leprosy cases, the PCR on SSS detected 23 (38.3%) in MB and 6 (9.9%) in PB forms. The ZN staining was picked 21 (35%) in MB and 3 (5%) in PB cases and MB cases by H&E 32 (53.3%) and PB cases 13 (21.6%) was detected. As to FF staining, 30 (50 %) in MB and 7 (11.6%) in PB cases were detected. This is a comparison of different laboratory methods with clinical classification. The amplified PCR products for each sample were run on gel electrophoresis and got 29 (48.33%) samples being positive by PCR on SSS. Among these positive samples, 3 MB and 2 PB samples were from AFB negative slides. The figure below shows the 500bp bands in agarose gel for M. leprae detection.
Figure 1: PCR result of SSS samples showing M. leprae positive and negative samples. 1= 1kb DNA ladder; 2,3 and 5 Positive samples of patients; 4 and 6 Negative Sample of patients; 8-Negative control and 10-Positive control.
Discussion
Our study demonstrated a positive yield in 45% of all cases of clinically diagnosed leprosy cases by SSS PCR and SSS microscopy 40%. Although the difference between these methods is insignificant, the result from SSS PCR indicated that it is possible to detect cases not picked by the ZN staining but needs certain improvements to exploit the potential of this advanced technique. The possibility of extracting DNA and running PCR from ZN-stained skin smear slides has an added advantage in the screening of smear-negative slides in leprosy, which will contribute to accuracy in the diagnosis and treatment of leprosy. Many studies have reported the advantages of PCR to detect M. leprae compared to SSS microscopy [5].
Conclusion
The number of positive results by H&E staining is higher than SSS PCR and this is mainly because of the nature of the disease. M. leprae is an intracellular bacterium that primarily resides in macrophages, these skin macrophages are found in the dermis part of the skin. Hence, introducing molecular diagnosis mainly PCR in addition to AFB staining of skin slit smear and histopathology confirmation of suspected leprosy cases is important for a definite diagnosis. The positive PCR result on stained skin slit smear was low in our study when it is compared with other studies. Therefore, there is a need for further improvement of PCR on SSS samples. This will contribute to simplifying the diagnostic process by avoiding invasive procedures. It also contributes to detecting patients with a low bacillary load as they are difficult to clearly define clinically. Recently, the GeneXpert is being used for TB diagnosis with the aim of replacing the routine AFB staining. Similarly, if we collect pieces of evidence on the potential of PCR in diagnosing leprosy, there will be a possibility of integrating the diagnosis for TB and leprosy to efficiently use the available resource throughout the country.
References
- P Singh, ST Cole. (2011). Mycobacterium Leprae: Genes, Pseudogenes and Genetic Diversity. Future Microbiol. 6(1):57-71.
Publisher | Google Scholor - RO Pinheiro, J de Souza Salles, EN Sarno, EP Sampaio. (2011). Sampaio Mycobacterium Leprae–Host-Cell Interactions and Genetic Determinants in Leprosy an Overview. Future microbiology. 6(2):217-230.
Publisher | Google Scholor - Noto S SP. (2011). Diagnosis of Leprosy. Leprosy Mailing List Archives.
Publisher | Google Scholor - Ruiz-Fuentes JL DA, Entenza AE, Frión Y, Suárez O, Torres P, et al. (2015). Comparison Of Four DNA Extraction Methods for The Detection of Mycobacterium Leprae from Ziehl–Neelsen-Stained Microscopic Slides. International Journal of Mycobacteriology. 4(4):284-289.
Publisher | Google Scholor - Santos AR DMA, Sarno EN, Suffys PN, Degrave WM. (1993). Use Of PCR-Mediated Amplification of Mycobacterium Leprae DNA in Different Types of Clinical Samples for The Diagnosis of Leprosy. J Med Microbiol. 39:298-304.
Publisher | Google Scholor