Research Article
Investigating the Prevalence of Clarithromycin Resistance Among Helicobacter Pylori Strains Isolated from Patients with Digestive Disorder
- Alireza Ahmadzadeh 1*
- Zhaleh Mohsenifar 2
- Behzad Hatami 3
- Ali Pirsalehi 4
- Ali Pirsalehi 5
- Mostafa Rezaei-Tavirani 6
- Zobayde Ahmadzadeh 7
Department of lablatory Sciences, Faculty of Paramedical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
*Corresponding Author: Alireza Ahmadzadeh, Department of lablatory Sciences, Faculty of Paramedical Sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Citation: Ahmadzadeh A, Mohsenifar Z, Hatami B, Pirsalehi A, Faeghi F, et al. (2024). Investigating the Prevalence of Clarithromycin Resistance among Helicobacter Pylori Strains Isolated from Patients with Digestive Disorder. Journal of Internal and Clinical Medicine, BioRes Scientia Publishers. 1(1):1-7. DOI: 10.59657/jicm.brs.24.004
Copyright: © 2024 Alireza Ahmadzadeh, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: April 10, 2024 | Accepted: May 01, 2024 | Published: July 08, 2024
Abstract
Introduction: Rise of resistant to clarithromycin which is a crucial component for eradication of Helicobacter pylori is a global concern. Point mutations in 23SrRNA gene is one of the most reason of clarithromycin resistance. This research aimed to explore the frequency of clarithromycin resistance and its connection with 23SrRNA point mutation.
Methods: This study was done on 100 patients who had referred to Valiasr Hospital suffering from gastric disorder during 2022. Two biopsy samples were taken from each patient and used for pathological and microbiological examinations. Antimicrobial vulnerability tests were performed by agar dilution and PCR.
Results: Out of the patients 53% (53/100) were diagnosed as H. pylori+. Pathological outcomes demonstrated that 54.7% (29/53) of the H Pylori+ patients suffered from chronic Gastritis, 37.7% (20/53) from Sever Active Gastritis, and 7.5 % (4/53) with Intestinal Metaplasia.Clarithromycin resistant were found among 13.2% (7/53) of patients. The MIC values of 0.125 mg/L and 2 mg/L were determined as the MIC50 and MIC90 values, respectively. According to the PCR results, A2142G point mutations in 23SrRNA gene detected among all clarithromycin resistant strains.
Conclusion: Our findings shown the existence of 23S rRNA point mutations may be related with resistance to clarithromycin in H. pylori strains.
Keywords: helicobacter pylori; clarithromycin resistance; histopathological changes; minimum inhibitory concentration
Introduction
The World Health Organization International Agency for Research on Cancer (WHO-IARC) has categorized Helicobacter pylori as a type I carcinogen. H. pylori infection is a main risk factor for gastric cancer (GC) which is the third deriving cause of cancer-related death within the world [1]. Beside this, H. pylori can lead to peptic ulcer, gastritis and other gastrointestinal disorder [2]. H pylori infection extended throughout the world; “In 2015, it was estimated that more than half of the world's population infected by it [3]. This infection is more common in developing countries [4]. H. pylori contamination among the Iranian population is high and the age of acquisition of infection is low [5]. Because of various complication associated with H. pylori, eradication is the first choice for treatment. There are several drug regimens for the cure, and clarithromycin is a key component of some regime [6,7]. Clarithromycin as a macrolide antibiotic is effective antimicrobials agent against H. pylori. It stops protein synthesis via binding to the 50S bacterial ribosomal subunit [8]. unfortunately, these days resistance to Clarithromycin has been expanding. This resistance mostly occurs via mutation at A2143G, followed by A2142G and A2142C positions of 23sRNA gene [9, 10]. Due to the wide ranging of infection, permanent monitoring of H. pylori prevalence and it resistance to antimicrobial agent is required [11]. The main goal of this study is to survey prevalence of H.pylori resistant to Clarithromycin among Iranian patient with gastrointestinal disorder via serial dilution Sensitivity test and PCR method.
Materials and methods
Patient samples
This study was conducted in Iran at Valiasr Hospital in 2022. It included 100 patients with dyspepsia who had underwent endoscopy. The patients excluded who had recently taken medication. Demographic data were documented in a standard questionnaire form. Two gastric biopsies were taken from the antrum of each patient; one of the biopsies for isolation of the H. pylori strains and other one sent to pathological lab for examination of histological changes.
H. pylori culture
Culture of the biopsies was done by smearing the specimens on the surface of supplemented Brucella agar medium with lysed horse blood and antibiotics (Skirrow's supplement, containing vancomycin, trimethoprim, and polymyxin B). The plates were incubated at 37 °C under microaerobic conditions (85% N2, 10% CO2, 5% O2) for 4 to 7 days. In continue, suspected colonies were identified by cell morphology, Gram staining, urease, oxidase, catalase assays and PCR for H. pylori using glmM primers, as described before [12].
MICs Determination
Vulnerability of the strains to the clarithromycin was identified by the agar dilution method (Merck, CAS-Number 81103-11-9) according the last guideline of European Committee on Antimicrobial Susceptibility Testing (EUCAST)[13]. Different quantities of clarithromycin, at final concentration of 0.06 to 16 mg/L,were added to Mueller-Hinton agar medium (sigma Aldrich, CAS-Number 70191) containing 10 percentage defibrinated horse blood. H Pylori suspensions in sterile saline with a density of 3 McFarland scale, i.e., 108 cells (CFU)/1 ml were used for sensitive testing. The minimal inhibition concentration (MIC) was determined as the lowest concentration of antibiotic that prevented the growth of the bacteria after 72 h of incubation at 37 °C under microaerophilic conditions [14]. H Pylori strains were considered to be vulnerable for MIC if MIC ≤ 0.25 mg/L, intermediate (MIC 0.5 mg/L), and resistant (MIC > 0.5 mg/L).
Molecular identification and mutation analysis via PCR
Fresh colonies of the H. pylori were utilized for DNA extraction using DNA extraction mini kit (YTA Genomic DNA Extraction Mini Kit for Tissue, Yektatajhiz, Tehran, Iran). DNA extracts were stored at –20 °C for further analysis. To identify H Pylori strains at species level PCR reaction was performed for glmM and 16S rRNA. In order to detect mutations of 23S rRNA at A2143G, A2142G positions, a volume of 25-μL reaction mixture which containing:1x PCR buffer, 0.3 μM of each primer, 1 μL of genomic DNA, 200 μM of dNTPs mix, 0.63 mmol of MgCl2, and 0.2 U/μL of Taq DNA polymerase was used. PCR reactions were done through Eppendorf thermal cycler (Germany; AG 22331) under the following conditions: 1 cycle of denaturation for 5 min at 94 °C, annealing for 5 min at 36 °C, extension for 5 min at 72 °C, followed by 30 cycles of denaturation for 1 min at 94 °C, annealing for1 min at 36 °C, and extension for 2 min at 72 °C. The PCR products were runded on electrophoresis in 1.2% agarose gel and stained by ethidium bromide. Primer sets which utilized in this research and PCR products size are shown in Table 1. DNA extracts of three reference strains encoding A2142G, A2143G (accession numbers JQ765438, JQ765441,) were used as control strains.
Table 1: Primers sequences and PCR products size used in this study
| Primer name | Primer sequence 5′→3′ | Product size | Reference |
| 16S rRNA | F: GGCTATGACGGGTATCCGGC | 764 | (15) |
| R: GCCGTGCAGCACCTGTTTTC | |||
| glmM | F: GGATAAGCTTTTAGGGGTGTTAGGGG | 296 | (16) |
| R: GCTTACTTTCTAACACTAACGCGC | |||
| A2142G | F: ACGGCGGCCGTAACTATA | 175 | (17) |
| R: AGGTCCACGGGGTCTTC | |||
| A2143G | F: TCGAAGGTTAAGAGGATGCGTCAGTC | 118 | (18) |
| R: CCGCGGCAAGACAGAGA |
Ethics
This research has been given ethics approval code IR.SBMU.RETECH.REC.1400.1189 from Shahid Beheshti University of Medical Sciences. Informed consent forms according to protocols approved by the ethical review committee at Shahid Beheshti University of Medical Sciences were gotten from the patients.
Statistical analysis
Data and results were analysed with SPSS 22 and Graph- Pad Prism 6 software. Statistical analysis was performed using Chi-square and Fisher's exact test. p <0>
Results
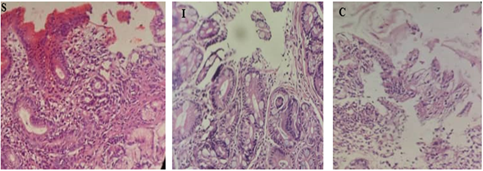
Out of 100 examined patients, 53 were contaminated with H. pylori. The age range of the infected patients was 18 - 60 years. among them, 32 were male (60.3%) and 21 were female (39.6%). based on pathological finding patients were divided into three group; 29(54.7%) of the patients were diagnosed with a chronic Gastritis (CG), 20 (37.7%) Sever Active Gastritis (SAG), and 4(7.5 %) with Intestinal Metaplasia (IM), Figure1.
Figure 1: pathological feature of H. pylori infected patient; S Sever Active Gastritis, I Intestinal Metaplasia, C chronic Gastritis.
According to demographic data analysis related to alcohol consuming, significant difference was found between CG and IM group (p< 0>H. pylori strains were resistant to clarithromycin, whereas 39 (73.5%) strains shown proneness. One (1.8%) strain was identified as intermediate. The MIC value of 4 mg/L was discovered as the most frequent among the resistant strains. The MIC values of 0.125 mg/L and 2 mg/L were confirmed as the MIC50 and MIC90 values, respectively. According to the MIC value H pylori resistant strain are located in IM group (57.1%),28.5% in SAG and 14.2% in CG respectively.100 % sensitivity or intermediate strain are stand in CG group.
Table 2: Demographic parameters of H. pylori-infected patients against pathological finding
| Parameter | Pathological Finding n=53 | |||
| n | CG | SAG | IM | |
| n=29 | n=20 | n=4 | ||
| Male | n=32 | 18(56.2%) | 11(34.3%) | 3(9.3%) |
| Female | n=21 | 11(52.3%) | 9(42.8%) | 1(4.7%) |
| Age | M | 18-45 | 35-59 | 55-60 |
| F | 22-48 | 30-57 | 52 | |
| smoking | M(n=7) | 4(57.1%) | 1(14.2%) | 2(28.5%) |
| F(n=2) | 1(50%) | 0(0%) | 1(50%) | |
| Alcohol consumption | M(n=6) | 1(16.6%) | 2(33.3%) | 3(50%) |
| F(n=1) | 0(0%) | 0(0%) | 1(100%) | |
| Blood group | M(n=32) | A (n=7, (38.8%)) | A (n=7, (63.3%)) | A (n=2, |
| B (n=5, (27.7%)) | AB (n=3, (27.2%)) | |||
| O (n=4, (22.2%)) | 0(n=1, (9%)) | (66.6%)) | ||
| Unknown (n=2, 11.1%)) | B (n=1, (33.3%)) | |||
| F(n=21) | A (n=5, (45.4%)) | B (n=3, (33.3%)) | A (n=1, (100%)) | |
| 0(n=6, (54.5%)) | O (n=5, (55.5%)) | |||
| Unknown (n=1, (11,1%)) | ||||
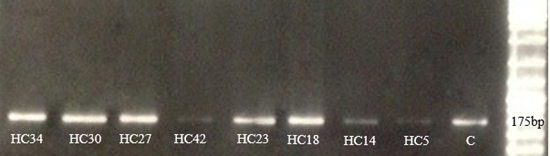
In order to elucidate the mechanism of clarithromycin resistance in H pylori isolate PCR was done with specific primer sets (Table 1) for detecting point mutations in 23srRNA. The A2142G point mutation was identified in 8/53 (~15%) specimens, whereas the A2143G mutation was not detected in samples. The strains that were identified by PCR are the ones that were previously identified by the MIC Figure 2. according to the pathological result, H pylori isolate HC34, HC30, HC27, HC42 are placed in Intestinal Metaplasia (IM) group HC23, HC18 in sever active gastritis (SAG) and HC14 in chronic Gastritis (CG) respectively. HC5 is related to the strain that has relative sensitivity or intermediate to clarithromycin and is placed in CG group. According to clarithromycin resistance rates and pathological findings considerable differences were found between IM and CG groups (p<0>
Table 3: MIC amount for clarithromycin among H. pylori isolates in different pathological finding
| MIC (mg/L) | Gender | pathological finding | ||
| n=53 | CG | SAG | IM | |
| n=29 | n=20 | n=4 | ||
| 0.06 | M(n=11) | 3(27.2%) | 8(72.7%) | 0 |
| F(n=10) | 6(60%) | 4(40%) | 0 | |
| 0.125 | M(n=8) | 7(87.5%) | 1(12.5%) | 0 |
| F(n=5) | 2(40%) | 3(60%) | 0 | |
| 0.25 | M(n=8) | 7(87.5%) | 1(12.5%) | 0 |
| F(n=3) | 2(66.6%) | 1(33.3%) | 0 | |
| 0.5 | M(n=0) | 0 | 0 | 0 |
| F(n=1) | 1(100%) | 0 | 0 | |
| 1 | M(n=1) | 0 | 1(100%) | 0 |
| F(n=0) | 0 | 0 | 0 | |
| 2 | M(n=0) | 0 | 0 | 0 |
| F(n=1) | 0 | 1(100%) | 0 | |
| 4 | M(n=3) | 1(33.3%) | 0 | 2(66.6%) |
| F(n=0) | 0 | 0 | 0 | |
| 8 | M(n=0) | 0 | 0 | 0 |
| F(n=1) | 0 | 0 | 1(100%) | |
| 16 | M(n=1) | 0 | 0 | 1(100%) |
| F(n=0) | 0 | 0 | 0 | |
CG chronic Gastritis, SAG sever active gastritis, IM Intestinal Metaplasia, M male, F female,MIC minimum inhibitory concentration
Figure 2: PCR amplification for the A2142G point mutation in 23S rRNA genes.
Discussion
H. pylori infection is the most predominant chronic bacterial disease that affecting nearly 50% of the world’s population; more than 80% contamination rats has been reported in developing countries as well [19]. This research reported 53 % infection rate which is similar to previous studies in Iran that have been described infection rate between 36% - 90% [20]. Administration of effective drug regimens in order to achieving high eradication rates of H pylori, is an important factor. The first-line drug regimen which includes a proton pump inhibitor (PPI), clarithromycin and amoxicillin or metronidazole, is used to treat H. pylori infection as standard triple therapy.So, resistance or sensitivity to Clarithromycin is one of the most important factors determining success or failure of treatment [21]. the occurrence of H. pylori resistance to clarithromycin is varies in different geographical region. Although this frequency is lower in developed countries, it is higher in developing countries, extending from 25% to 50%. [22] A total of 21 studies on clarithromycin resistance of H. pylori in different parts of Iran were conducted; the range of clarithromycin resistant is reported to be between highest (75%) and the lowest (0%) [23]. Another study in Iran reported MIC values of 0.25 mg/L and 16 mg/L as the MIC50- MIC90 respectively [24]. According to our study, MIC value and PCR approval showed13.2% of H. pylori strains were resistant to clarithromycin and 0.125 mg/L, 2 mg/L were determined as the MIC50 and MIC90 values, respectively. The same investigate in china has been stated MIC50 0.0312mg/L, MIC90 64mg/L [25]. A survey in 24 centres from 18 European countries have been reported clarithromycin resistance range 4.8–36.9 % [26]. look like this research in Italy, 13.5% resistance to clarithromycin were found among Helicobacter pylori strains [27]. Clarithromycin works through interaction with the peptidyl transferase domain V of the 23S rRNA subunit; hence 23S rRNA Point mutations have been found to cause decreasing clarithromycin affinity and resulting in bacterial resistance to the antibiotic. A2142G, A2143G, are the most prevalent point mutations [28]. in a recent study conducted by Farzi et al from Iran, among 23 clarithromycin resistance strain just 17.3 % (4/23) recognized with A2143G point mutation. In contrast, Khashei et al. reported that A2142G was the most (90%) frequent point mutation [29]. in a study in Korea, A2143G was the most (19.5%), significant point mutation and only 0.9% was reported as A2142G [30]. However current research detected 100 % of clarithromycin resistance strain carry the A2142G point mutation.
Conclusions
In conclusion, this study revealed that the occurrence of H. pylori clarithromycin resistance is expanding in Iran. The findings from this study also highlight the relevance of types of mutations in genes responsible for antibiotic resistance in H. pylori strains. We also provide evidence for the importance of screening of resistance genotypes in H. pylori strains for guiding clinicians to choose an appropriate combination of drugs.
Acknowledgements
Thanks to all who contributed to this study. All authors approved the final version of the manuscript
Abbreviation
H Pylori; Helicobacter pylori
CG; Chronic Gastritis
SAG; Sever Active Gastritis
IM; Intestinal Metaplasia
MIC; Minimal Inhibition Concentration
References
- Guo Y, Cao XS, Guo GY, Zhou MG, Yu B. (2022). Effect of Helicobacter Pylori Eradication on Human Gastric Microbiota: A Systematic Review and Meta-Analysis. Front Cell Infect Microbiol., 12:899248.
Publisher | Google Scholor - Cho JH, Jin SY. (2022). Current guidelines for Helicobacter pylori treatment in East Asia 2022: Differences among China, Japan, and South Korea. World J Clin Cases., 10(19): 6349-6359
Publisher | Google Scholor - Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, et al. (2017). Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology.,153(2):420-9.
Publisher | Google Scholor - Tran V, Saad T, Tesfaye M, Walelign S, Wordofa M, Abera D, et al. (2022). Helicobacter pylori (H. pylori) risk factor analysis and prevalence prediction: a machine learning-based approach. BMC Infectious Diseases., 22(1):655.
Publisher | Google Scholor - Massarrat S, Saberi-Firoozi M, Soleimani A, et al. (1995). Peptic ulcer disease, irritable bowel syndrome and constipation in two populations in Iran. Eur J Gastroenterol Hepatol., 7:427–33.
Publisher | Google Scholor - Goldberg L, Amrick TJ. (2022). Successful Eradication of Helicobacter pylori with 5-Day Concomitant Treatment. GastroHep., 2022:1211329.
Publisher | Google Scholor - Leung WK, Graham DY. (2000). Clarithromycin for Helicobacter pylori infection. Expert Opin Pharmacother., 1(3):507-14.
Publisher | Google Scholor - Kocsmár É, Buzás GM, Szirtes I, Kocsmár I, Kramer Z, Szijártó A, et al. (2021). Primary and secondary clarithromycin resistance in Helicobacter pylori and mathematical modeling of the role of macrolides. Nature Communications., 12(1):2255.
Publisher | Google Scholor - Hosseini RS, Rahimian G, Shafigh MH, Validi M, Khaledi M, Gholipour A. (2021). Correlation between clarithromycin resistance, virulence factors and clinical characteristics of the disease in Helicobacter pylori infected patients in Shahrekord, Southwest Iran. AMB Express., 11(1):147.
Publisher | Google Scholor - Albasha AM, Elnosh MM, Osman EH, Zeinalabdin DM, Fadl AAM, Ali MA, et al. (2021). Helicobacter pylori 23S rRNA gene A2142G, A2143G, T2182C, and C2195T mutations associated with clarithromycin resistance detected in Sudanese patients. BMC Microbiology., 21(1):38.
Publisher | Google Scholor - Bordin D, Morozov S, Plavnik R, Bakulina N, Voynovan I, Skibo I, et al. (2022). Helicobacter pylori infection prevalence in ambulatory settings in 2017-2019 in RUSSIA: The data of real-world national multicenter trial. Helicobacter., e12924.
Publisher | Google Scholor - Gharibi S, Falsafi T, Alebouyeh M, Farzi N, Vaziri F, Zali MR. (2017). Relationship between histopathological status of the Helicobacter pylori infected patients and proteases of H. pylori in isolates carrying diverse virulence genotypes. Microbial Pathogenesis., 110:100-6.
Publisher | Google Scholor - Kahlmeter G, Brown DF, Goldstein FW, et al. (2006). European Committee on Antimicrobial Susceptibility Testing (EUCAST) Technical Notes on antimicrobial susceptibility testing. Clin Microbiol Infect.,12(6):501-503.
Publisher | Google Scholor - Bińkowska A, Biernat MM, Łaczmański Ł, Gościniak G. (2018). Molecular Patterns of Resistance Among Helicobacter Pylori Strains in South-Western Poland. Front Microbiol., 9:3154. Published 2018 Dec 18.
Publisher | Google Scholor - Bohr UR, Primus A, Zagoura A, Glasbrenner B, Wex T, Malfertheiner P. (2002). A group-specific PCR assay for the detection of Helicobacteraceae in human gut. Helicobacter., 7(6):378-383.
Publisher | Google Scholor - Kauser F, Hussain MA, Ahmed I, et al. (2005). Comparative genomics of Helicobacter pylori isolates recovered from ulcer disease patients in England. BMC Microbiol., 5:32.
Publisher | Google Scholor - Pan ZJ, Su WW, Tytgat GN, Dankert J, van der Ende A. (2002). Assessment of clarithromycin-resistant Helicobacter pylori among patients in Shanghai and Guangzhou, China, by primer-mismatch PCR. J Clin Microbiol., 40(1):259-261.
Publisher | Google Scholor - Furuta T, Soya Y, Sugimoto M, et al. (2007). Modified allele-specific primer-polymerase chain reaction method for analysis of susceptibility of Helicobacter pylori strains to clarithromycin. J Gastroenterol Hepatol., 22(11):1810-1815.
Publisher | Google Scholor - Alexander SM, Retnakumar RJ, Chouhan D, Devi TNB, Dharmaseelan S, Devadas K, et al. (2021). Helicobacter pylori in Human Stomach: The Inconsistencies in Clinical Outcomes and the Probable Causes. Frontiers in Microbiology, 12.
Publisher | Google Scholor - Fakheri H, Saberi Firoozi M, Bari Z. (2018). Eradication of Helicobacter Pylori in Iran: A Review. Middle East J Dig Dis, 10(1):5-17.
Publisher | Google Scholor - Kim SY, Chung JW. (2020). Best Helicobacter pylori Eradication Strategy in the Era of Antibiotic Resistance. Antibiotics (Basel), 9(8):436.
Publisher | Google Scholor - Yilmaz O, Demiray E. (2007). Clinical role and importance of fluorescence in situ hybridization method in diagnosis of Helicobacter pylori infection and determination of clarithromycin resistance in H. pylori eradication therapy. World. J. Gastroenterol, 13:671-675
Publisher | Google Scholor - Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG. (2015). Helicobacter pylori in Iran: A systematic review on the antibiotic resistance. Iran J Basic Med Sci, 18(1):2-7.
Publisher | Google Scholor - Alavifard H, Mirzaei N, Yadegar A, et al. (2021). Investigation of Clarithromycin Resistance-Associated Mutations and Virulence Genotypes of Helicobacter pylori Isolated from Iranian Population: A Cross-Sectional Study. Curr Microbiol., 78(1):244-254.
Publisher | Google Scholor - Huang X, Liu Y, Lin Z, et al. (2021). Minimum inhibitory concentrations of commonly used antibiotics against Helicobacter Pylori: A multicenter study in South China. PLoS One, 16(9): e0256225.
Publisher | Google Scholor - Megraud F, Bruyndonckx R, Coenen S, et al. (2021). Helicobacter pylori resistance to antibiotics in Europein 2018 and its relationship to antibiotic consumption in the community. Gut, 70:1815–1822.
Publisher | Google Scholor - De Francesco V, Zullo A, Fiorini G, Saracino IM, Pavoni M, Vaira D. (2018). Role of MIC levels of resistance to clarithromycin and metronidazole in Helicobacter pylori eradication. Journal of Antimicrobial Chemotherapy, 74(3):772-4.
Publisher | Google Scholor - Hussein RA, Al-Ouqaili MTS, Majeed YH. (2022). Detection of clarithromycin resistance and 23SrRNA point mutations in clinical isolates of Helicobacter pylori isolates: Phenotypic and molecular methods. Saudi J Biol Sci, 29(1):513-520.
Publisher | Google Scholor - Khashei, R.; Dara, M.; Bazargani, A.; Bagheri Lankarani, K.; Taghavi, A.; Moeini, M.; Dehghani, B.; Sohrabi, M. (2016). High rate of A2142G point mutation associated with clarithromycin resistance among Iranian Helicobacter pylori clinical isolates. Apmis Acta Pathol. Microbiol. Immunol. Scand, 124, 787–793.
Publisher | Google Scholor - Seo SI, Do BJ, Kang JG, Kim HS, Jang MK, Kim HY, Shin WG. (2020). Helicobacter pylori Eradication According to Sequencing-Based 23S Ribosomal RNA Point Mutation Associated with Clarithromycin Resistance. Journal of Clinical Medicine, 9(1):54.
Publisher | Google Scholor