Editorial letter
Decoding the Genomic Complexity of Sepsis: Paving the Way for Future Advances
1Genetics department, Bhagwan Mahavir Medical Research Centre, Hyderabad, India.
2Medical Director, Mahavir Hospital and Research Centre, Hyderabad, India.
*Corresponding Author: Kaiser Jamil, Genetics department, Bhagwan Mahavir Medical Research Centre, Hyderabad, India.
Citation: Jamil K, Iyengar S. (2023). Decoding the Genomic Complexity of Sepsis: Paving the Way for Future Advances. Clinical Case Reports and Studies, BioRes Scientia Publishers. 3(6):1-4. DOI: 10.59657/2837-2565.brs.23.091
Copyright: © 2023 Kaiser Jamil, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: December 02, 2023 | Accepted: December 18, 2023 | Published: December 25, 2023
Abstract
Sepsis, a life-threatening condition resulting from the body's extreme response to infection, remains a global health challenge with high morbidity and mortality rates. The primary aim of this article is to unravel the intricate genetic factors contributing to sepsis susceptibility, severity, and patient outcomes. By understanding the genomic landscape of sepsis and its pathogenic infiltrations, this article tries to contribute to the development of novel diagnostic tools and therapeutic interventions, additionally it tries to integrate genomic data with clinical information to develop personalized approaches for sepsis management, including targeted therapies, drug development and precision medicine, and improving overall outcomes for individuals affected by Sepsis.
Keywords: sepsis; genetics; snps; pathogens; immune response; organ failure; meta-analysis; ngs
Introduction
Sepsis stands as a formidable challenger within the field of medicine. It lurks in the shadows, often striking swiftly and mercilessly, claiming lives with alarming efficiency. The fight against sepsis has been ongoing for centuries, but it is only in recent years that we have begun to uncover the intricate genetic underpinnings of this deadly condition [1, 2, 3]. As we delve deeper into the genetics of sepsis, a new dawn of hope emerges on the horizon. Sepsis is not a single disease but rather a complex syndrome triggered by the body's exaggerated response to infection. It can result from many infections, including bacterial, viral, and fungal. Its symptoms can range from fever and organ dysfunction to septic shock and death [4,5]. Despite advances in medical science, sepsis remains a leading cause of morbidity and mortality worldwide, with an estimated 20 million cases and 11 million deaths annually as infectious organisms such as bacteria, viruses, and fungi become resistant to the antibiotic- drugs used to treat them.
Genetics has long been recognized as a contributing factor to the risk and severity of sepsis [6]. The hereditary component of sepsis susceptibility has been a topic of interest for researchers for years, and recent advancements in genetic studies have brought us closer to understanding the genetic determinants of this condition. One of the key findings in the genetics of sepsis is the identification of specific genetic variations that increase an individual's susceptibility to the syndrome [2]. These variations can affect various components of the immune system, making some people more vulnerable to overwhelming infections. For example, gene variations related to pattern recognition receptors (PRRs), which play a crucial role in detecting pathogens, can influence how an individual responds to infection. In silico studies or using bioinformatics can shorten our list of therapeutic options [7]. Understanding these genetic predispositions can help us identify individuals at higher risk and potentially develop personalized treatment strategies.
NGS techniques like whole-genome sequencing (WGS) and whole-exome sequencing (WES) allow researchers to examine an individual's entire genome or just the protein-coding regions (exome) to identify rare genetic variants associated with sepsis. Functional genomics studies aim to understand the biological consequences of genetic variations. This involves investigating how specific genetic variants affect gene expression, protein function, or immune response pathways related to sepsis. Meta-analysis combines data from multiple genetic studies to increase statistical power and identify more robust genetic associations with sepsis [8,9]. These analytical methods are potent tools that can conquer the complications of large-scale randomized controlled trials.
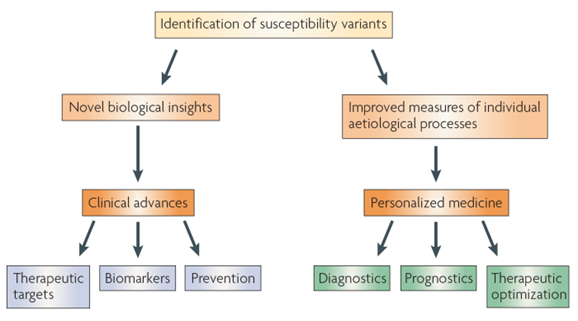
The genetic predisposition to inflammation, it is observed that genetic variations in pro-inflammatory and anti-inflammatory cytokines, such as interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α), can impact an individual's response to infection in sepsis cases. Polymorphisms in these genes have been linked to variations in the severity of sepsis, such as sepsis, severe sepsis and septic shock, considering patient outcomes [10]. The figure-1 summarizes the steps to identify the genetic variants in diseases.
Figure 1
Source: Google images from Nature
Furthermore, the genetic basis of sepsis severity has recently also come into focus; this predicts that certain genetic factors can influence the body's response to infection, leading to a dysregulated immune response and organ dysfunction. Various authors have also listed a genetic variation in several other genes. Targeting these genetic markers could offer new avenues for intervention and treatment. This field, known as pharmacogenomics, most commonly reported in human cancers, studies how an individual's genetic makeup influences their response to medications; this is another promising area in sepsis research [11]. Tailoring drug treatments based on a patient's genetic profile could enhance the effectiveness of therapies and reduce the risk of adverse events. This approach can potentially revolutionise sepsis management, as it could lead to more personalized and precise treatments. However, the other face of this multi-factorial investigation is the study of the genetics of the specific pathogen and various sources of drugs present in the armamentarium. This would redesign the costly affair of inventing new molecules to treat sepsis.
In sepsis the concept of tailored drugs may be new, and also known as personalized or precision medicine, involves customizing medical treatment to the individual characteristics of each patient. In the context of infections and drug-resistant pathogens, the idea is to design drugs that specifically target the unique features of a particular strain of the pathogen affecting an individual. In this regard we have several options for overcoming the challenges posed by multiple drug-resistant strains of pathogens: firstly, tailored drugs can be designed to target specific molecular structures or pathways that are essential for the survival of the resistant pathogen. Secondly, using genomic analysis technology the genetic makeup of the pathogen could be analyzed. By understanding the genetic variations among different strains, their exists an immense opportunity to determine the vulnerabilities which can be exploited to design drugs with high specificity and efficacy. Also, the benefit of such tailored drugs may have fewer side effects compared to broad-spectrum antibiotics or antivirals. Since they are designed to act specifically on the target pathogen, they may spare the beneficial microbes in the body, reducing the risk of collateral damage and associated complications. Immunotherapies are one more option, combining different strategies can provide a multi-pronged attack on the pathogen.
While the concept of tailored drugs holds great promise, there are challenges, including the need for advanced diagnostic tools, cost considerations, and regulatory hurdles. Additionally, the rapid evolution of pathogens requires continuous research and development to stay ahead of emerging drug-resistant strains. Nonetheless, personalized medicine, including tailored drugs, represents a significant step toward more effective and efficient treatments for not only Sepsis, but also for other infectious diseases. An Abrader-based pharmacogenomic type approach to sepsis would be the new wave of research which can help humanity.
However, while the genetics of sepsis hold great promise, it also presents challenges. The genetic factors involved in sepsis are complex and multifaceted, making it difficult to pinpoint specific genes or mutations responsible for the condition. Additionally, genetic susceptibility, primarily investigated in diabetes, cancers and cardiovascular diseases, is just one piece of the puzzle. It has yet to be investigated thoroughly in sepsis, as sepsis results from a complex interplay between genetics, environmental factors, and the Nature of the infecting pathogens [12].
Conclusion
In summary, the study of the genetics of sepsis is a beacon of hope in our quest to combat this devastating condition. While we may have some of the answers, the progress made in recent years is remarkable. By unravelling the genetic tapestry of sepsis, we inch closer to more effective prevention, early detection, and personalized treatment strategies. This journey is a testament to the power of science and human determination, and it offers a glimmer of hope to the countless individuals and families affected by sepsis worldwide [13]. As genetic research advances, we can anticipate more precise risk stratification, targeted therapies, and improved outcomes for sepsis patients based on their genetic profiles. However, further research and larger-scale studies are needed to validate these findings and integrate genetics into routine clinical practice. As we continue deciphering the genetic code of sepsis, let us remain steadfast in our commitment to turning the tide against this formidable foe.
Declarations
Acknowledgement
We are thankful to the Chairman and Research Director for their encouragement.
Conflict of Interest
Authors declare No Conflict of Interest
Disclosure
No funding was received for conducting this study.
Author Contributions
All authors contributed equally to this concept and design.
References
- Engoren M, Jewell ES, Douville N, Moser S, Maile MD, Bauer ME. (2022). Genetic variants associated with sepsis. PLoS One, 17(3):e0265052.
Publisher | Google Scholor - Giamarellos-Bourboulis EJ, Opal SM. The role of genetics and antibodies in sepsis. Ann Transl Med., 4(17):328.
Publisher | Google Scholor - Naga Chaitanya, Kaiser Jamil, and Mohammad Abdul Azeem Zahid (2023). CONFRONTING SEPSIS: A CALL FOR URGENT ACTION, European Journal of Pharmaceutical and Medical Research. Ejpmr, 10(7):200-202.
Publisher | Google Scholor - Villar, J., Maca-Meyer, N., Pérez-Méndez, L. et al. (2004). Bench-to-bedside review: Understanding genetic predisposition to sepsis. Crit Care, 8:180
Publisher | Google Scholor - MedlinePlus homepage, Bethesda (M.D.): National Library of Medicine (U.S.)
Publisher | Google Scholor - Abu-Maziad, A., Schaa, K., Bell, E. et al. (2010). Role of Polymorphic Variants as Genetic Modulators of Infection in Neonatal Sepsis. Pediatr Res, 68:323-329
Publisher | Google Scholor - Zeng X, Feng J, Yang Y, Zhao R, Yu Q, et al. (2021). Screening of Key Genes of Sepsis and Septic Shock Using Bioinformatics Analysis. J Inflamm Res., 14:829-841.
Publisher | Google Scholor - Rosier, F.; Brisebarre, A.; Dupuis, C.; Baaklini, S.; Puthier, D.; (2021). Genetic Predisposition to the Mortality in Septic Shock Patients: From GWAS to the Identification of a Regulatory Variant Modulating the Activity of a CISH Enhancer. Int. J. Mol. Sci., 22:5852.
Publisher | Google Scholor - D'Urso, S., Rajbhandari, D., Peach, E., De Guzman, E., Li, et al. (2020). Septic Shock: A Genomewide Association Study and Polygenic Risk Score Analysis. Twin Research and Human Genetics, 23(4):204-213.
Publisher | Google Scholor - J.M.Ferrer Agüeroa, S.Millánb,c, F.Rodríguezde Castrod, e, I.Martín-Loechesb,c, J. SoléViolán (2013). Community-acquired pneumonia: Genetic variants influencing systemic inflammation: Review. Med Intensiva, 38(5):315-323.
Publisher | Google Scholor - Zhang, A. Q., & Yue, C. L. (2014). Interleukin-10 gene polymorphisms and susceptibility to sepsis: a meta-analysis. Inflammation Research, 63(1):25-32.
Publisher | Google Scholor - Lu, H., Wen, D., Wang, X. et al. (2019). Host genetic variants in sepsis risk: a field synopsis and meta-analysis. Crit Care, 23:26.
Publisher | Google Scholor - Zehnbauer, B.A., Freeman, B.D., Buchman, T.G. (1999). Genetic Predisposition to Sepsis and Organ Failure. In: Baue, A.E., Berlot, G., Gullo, A., Vincent, JL. (eds) Sepsis and Organ Dysfunction. Springer, Milano.
Publisher | Google Scholor