Review Article
Base Excision Repair: A Review
- Ashwin Sidhu *
California Northstate University College of Medicine, Elk Grove, California, United States.
*Corresponding Author: Ashwin Sidhu, California Northstate University College of Medicine, Elk Grove, California, United States.
Citation: Ashwin Sidhu. (2023). Base Excision Repair: A Review. Journal of BioMed Research and Reports, BRS Publishers. 2(4):1-3; DOI: 10.59657/2837-4681.brs.23.032
Copyright: © 2023 Ashwin Sidhu, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: July 20, 2023 | Accepted: August 04, 2023 | Published: August 08, 2023
Abstract
Base excision repair (BER) is a fundamental DNA repair pathway essential for maintaining the genomic integrity of all living organisms. This paper provides an overview of the mechanisms, key players, and regulatory aspects of the base excision repair pathway. BER serves as the primary defense against spontaneous DNA damage arising from endogenous and exogenous sources, including oxidative stress, deamination, and alkylation. This repair process involves a sequential series of enzymatic reactions orchestrated by various proteins, including DNA glycosylases, AP endonucleases, DNA polymerases, and DNA ligases.
Keywords: excision; repair; human genome; mutagens; ultraviolet radiation
Introduction
The human genome is constantly exposed to a wide variety of mutagens whether it is alkylating agents, intercalating agents, ultraviolet radiation, or ionizing radiation. Each type of mutagen leads to DNA damage. For instance, the DNA damages associated with ultraviolet radiation are helix distorting and the formation of pyrimidine dimers, while alkylating agents add bulky sides group to DNA and change base pairing. With DNA damage comes various mechanisms of DNA repair. Mismatch repair, nonhomologous end-joining, and nucleotide excision repair are just a few mechanisms used to repair damaged DNA. One major repair pathway is base excision repair (BER), which eliminates single damaged-base residues [1].
Description
Base excision repair can be described as a highly regulated network of enzymes responsible for repair of modified base lesions [2]. Base excision repair is a DNA repair mechanism that corrects for errors caused by oxidation, alkylation, and deamination or base loss. BER is useful for correcting small base lesions and mostly focuses on small base lesions that cause very little distortion to the helix structure of DNA. Much of the damage that leads to the small base lesions arise from radiation or environmental chemicals. Specifically, BER responds to the damage caused by the deamination of cytosine and the formation of 8-oxoguanine formed from the oxidation of guanine [3].
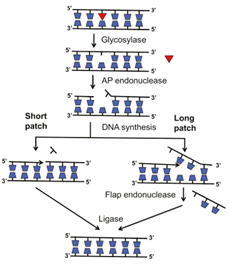
When uracil is being constantly formed by the deamination of cytosine, a UG mis-pair is generated. The uracil containing base is recognized and cleaved by the DNA uracil glycosylase. The cleavage of the uracil prevents transition mutations from occurring during the subsequent DNA replication. Thus, an excision of the uracil by the DNA uracil glycosylase results in an apyrimidinic site, which is a location in the DNA that does not has a pyrimidine base. Also, 8-oxoguanine is a major product of oxidation damage. Specifically, the exposure of guanine to H2O2 results in the formation of 8-oxoguanine. 8-oxoguanine is recognized and excised by DNA oxoguanine glycosylase, which leads to the formation of an apurinic site. Overall, DNA glycosylases are responsible for the initial recognition of the base lesion. Once the recognition is complete, the DNA glycosylases cleaves the N-glyosidic bond of the damaged base, which leaves an AP site [4]. After the DNA glycosylases recognize and cleave the damaged base, AP endonuclease excises the empty AP site in the DNA strand. When the AP endonuclease cleaves the baseless sugar from the DNA strand, a single-stranded gap is left. After AP endonuclease activity, an enzyme known as dRPase (DNA deoxy ribo phosphodiesterase) removes the sugar-phosphate residues from the duplex DNA [1]. For ligation of the bases to occur in base excision repair, end processing must occur. AP endonucleases and polynucleotide kinase-phosphatase participate in 3' end processing with PNKP promoting the hydroxyl group on the 3’ end and the phosphate group of the 5’ end. Again, 3’ end processing is necessary for DNA synthesis because DNA polymerase needs the 3’ hydroxyl group to extend the DNA strand. After end processing is complete, the gap is filled by DNA polymerase and the stand is ligated by DNA ligase. Specifically, DNA polymerase b is the polymerase that synthesizes the DNA strand during BER. There can be short-patch BER or long-patch BER, which involves the insertion of 2-10 nucleotides. In long-patch BER, a 5’ flap is generated, which is removed by flap endonuclease 1 (FEN1). In short-patch BER, FEN1 is not needed, and DNA ligase activity occurs directly after DNA polymerase b adds the new base to the DNA strand. The general schematic for base excision repair is shown below in figure 1 [5].
Figure 1: Basic schematic of Base Excision Repair [5].
As shown above, the major enzymes/proteins in base excision repair are DNA glycosylases, AP endonuclease, DNA polymerase b, flap endonuclease, and DNA ligase. If one of the enzymes failed to work properly, then base excision repair will not be completed and either a small base lesion or a single-stranded gap will be left in the DNA strand.
Various biological consequences and human disease arise from a defect in the base excision repair pathway. In fact, defects in base excision repair have links with cancer. For instance, deletion mutation in genes involved in base excision repair has shown to lead to higher mutation rates. In other words, the loss of base excision repair function contributes to cancer. Specifically, mutation in DNA polymerase b is found in about 30% of human cancers. In addition, mutations in DNA glycosylases also lead to cancer. Mutations in MYH, a specific type of DNA glycosylase, has shown to increase the susceptibility of colon cancer [6]. Mutations in other DNA glycosylases, such as MBD4 and NEIL1, also shows elevated cancer in humans. The mutations in MBD4 and NEIL1 are a result of epigenetic alterations. When MBD4 expression is reduced due to epimutations, the risk of colorectal neoplasms and cancers is increased. Furthermore, epigenetic mutations which reduces the expression of NEIL1 has shown to be a primary indicator of gastric cancer [6].
Conclusion
Overall, base excision repair is a complex network of enzymes that work together to repair a small base lesion. If one of the enzymes does not function properly, negative consequences may occur, and cancer is one of the diseases that can occur from a defect in base excision repair.
Conflicts of Interest
No conflicts of interest.
References
- Wood R.D. (1996). DNA Repair in Eukaryotes. Annu. Rev. Biochem, 65:137-144.
Publisher | Google Scholor - Schermerhorn K. M., Delaney, S. (2014). A Chemical and Kinetic Perspective on Base Excision Repair of DNA. Acc. Chem. Res, 47(4):1238-1246.
Publisher | Google Scholor - Hartl D. L., Cochrane, B. J. (2017). Genetics: Analysis of Genes and Genomes, 9th ed.; Jones & Bartlett Learning: Burlington, MA, 607-608.
Publisher | Google Scholor - Krokan, H. E., Bjoras, M. (2013). Base Excision Repair. Cold Spring Harbor Perspectives in Biology, 5:1-16.
Publisher | Google Scholor - Hinz, J. M., Czaja, W. (2015). Facilitation of base excision repair by chromatin remodeling. DNA Repair, 36:91-97.
Publisher | Google Scholor - Wallace S. S., Murphy D. L., Sweasy J. B. (2012). Base Excision Repair and Cancer. Cancer Lett, 327(1-2):73-89.
Publisher | Google Scholor