Review Article
Geospatial Analysis Applied to Epidemiological Studies of Rabies Disease: A Systematic Review
1Master in Tropical Health Innovation, Bobonaro Saude Municipal Service, Maliana City, Timor-Leste.
2Department of Biology, Faculty of Information, Technology and Science, Hindu University of Indonesia, Denpasar, Indonesia.
3Associate Epidemiologist, Indonesian Society of Epidemiologists, Special Capital Region of Jakarta 10560, Indonesia.
4One Health Laboratory Network (OHLN), Depok, West Java, 16424, Indonesia.
5Department of Epidemiological Surveillance, Bobonaro Saude Municipal Service, Maliana City, Timor-Leste.
*Corresponding Author: Zito Viegas da Cruz, Master in Tropical Health Innovation, Bobonaro Saude Municipal Service, Maliana City, Timor-Leste.
Citation: Zito V. Cruz, Adnyana M.D.M., de-Souza J. (2024). Geospatial Analysis Applied to Epidemiological Studies of Rabies Disease: A Systematic Review, Journal of BioMed Research and Reports, BioRes Scientia Publishers. 5(5):1-15. DOI: 10.59657/2837-4681.brs.24.114
Copyright: © 2024 Zito Viegas da Cruz, this is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Received: September 30, 2024 | Accepted: October 18, 2024 | Published: October 25, 2024
Abstract
Rabies is a deadly zoonotic disease that remains a global health problem. The incidence of this disease is increasing, but it has not yet been overcome by various parties. The easy transmission of this disease from animals to humans makes the selection of epidemiological analysis methods necessary to accelerate its eradication and control. This systematic review aimed to assess the types of spatial methods used in rabies epidemiological studies published between January 2014 and April 2024. Thirty-eight studies were selected, and 28 different spatial methods were used in rabies studies during that period, with two methods being the most frequently used. Few articles have applied spatial analysis methods in rabies studies; however, whenever they were applied, they contributed to a better understanding of the geospatial diffusion of rabies. This review highlights the importance of geospatial analysis in understanding the spread of rabies, identifying hotspots, and identifying the need for more targeted and effective interventions.
Keywords: analytical models; epidemiological studies; geospatial techniques; rabies; zoonotic disease
Introduction
Rabies is a deadly zoonotic disease that remains a global health concern, particularly in regions with low animal vaccination rates and limited access to medical care [1]. Although rabies can be prevented through vaccination and animal control measures, the disease remains widespread in many developing countries, causing thousands of deaths annually [2-5]. A major problem faced in rabies control is the lack of appropriate and efficient identification of patterns of disease spread.
The spread of rabies is influenced by a variety of environmental and social factors, including the distribution of reservoir animal populations, animal-human interactions, and geographical conditions that facilitate or inhibit disease spread [6]. Without an in-depth understanding of how these factors interact spatially, control efforts are likely ineffective. With the development of technology and science, new methods for analyzing and understanding the epidemiology of rabies have become critical for improving prevention and control strategies. Geographical factors and information from different sources and formats can be spatially combined by GIS, both in epidemiology and public health, as, for example, several previous studies have found that geographical factors include temperature, precipitation, and spatial distribution of rabies cases; risk estimation and spatial distribution mapping of rabies cases in different regions; predicting rabies distribution successfully; Spatial studies support rabies control practices and vaccination strategies [7-9]. Therefore, there is an urgent need for analytical methods that can integrate epidemiological and geospatial data to produce accurate risk maps and reliable prediction models.
This review is relevant in the context of global rabies control, particularly in endemic regions. By understanding how geospatial analysis can be used to map the spread of rabies, how effective it is in mapping the spread of rabies, identifying rabies hotspots, and identifying the challenges faced in applying geospatial analysis to rabies epidemiological studies, authorities can design more targeted and effective interventions. In addition, identifying rabies hotspots can facilitate ongoing surveillance and evaluation of the effectiveness of prevention programs. This approach can also be applied to other zoonotic diseases, making its contribution to global public health significant.
Systematizing knowledge on the application of geospatial analysis in rabies studies through a systematic review can provide comprehensive insight into existing approaches, their effectiveness, and research gaps that need to be filled. The aim of this review is to analyze the role of geospatial analysis in epidemiological studies, including its types, uses, and implications for society. The results of this systematic review will not only provide guidance for researchers and health practitioners in selecting and applying appropriate methods but also serve as a basis for the development of new tools and technologies for the mapping and control of complex zoonotic diseases.
Materials and Methods
Systematic Review Registration
This systematic review used the Preferred Reporting Items for Systematic Reviews and Meta-Analysis Protocol guidelines [10]. The PROSPERO registration number was added if applicable.
The Eligibility Criteria
At this stage, one author sorted and collected articles containing cross-sectional, cohort, observational, and experimental data. The eligibility of an article to be included in the study went through various stages and met the following established inclusion criteria: a) the article had to discuss spatial, spatial-temporal rabies, and spatial autocorrelation in rabies; b) it clearly presented spatial methods as part of data analysis and results preparation; c) it was published in the last decade (2014-2024); and d) it was listed in reputable journals indexed by PubMed, Scopus, and Web of Science (WOS). The range of years chosen depends on the novelty of the research, and the reputation of the journal determines the credibility of the article. Articles should contain methods, workflows, and supporting data that can be processed in spatial analysis and provide clear spatial outputs in accordance with the research objectives of mapping rabies disease. Detailed article eligibility must include data analysis units, typology representation, spatial methods used, and the main results of spatial data analysis. The exclusion criteria for the excluded articles included a) using other foreign languages, b) not open access, c) not only discussing rabies, d) discussing rabies but not using spatial analysis methods, and e) belonging to predatory publishers and journals. All articles that did not meet the eligibility criteria, were duplicates, were discontinued studies, or were irrelevant at this stage were excluded.
Sources of Information, Literature Search Strategies, and Study Selection
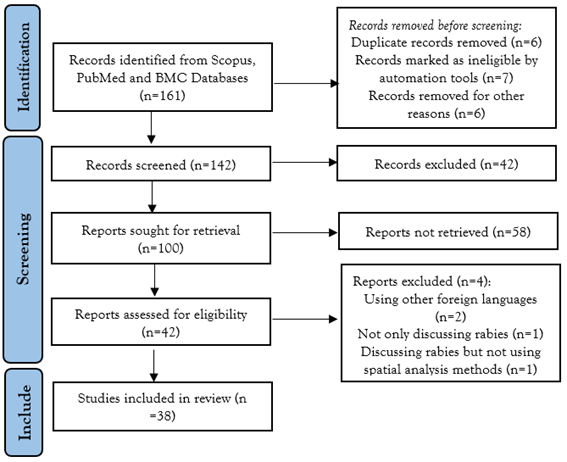
At this stage, one author collected and searched for appropriate literature by setting keywords including "spatiotemporal clustering of rabies,” "spatial clustering of rabies,” "spatial autocorrelation of rabies,” "rabies AND spatial,” "spatial-temporal AND rabies.” We used trusted databases and credible sources to search for articles that matched the topic in PubMed (MeSH terms). The PubMed (http://www.ncbi.nlm.nih.gov/PubMed) and BMC (https://www.biomedcentral.com) databases were searched. The collected articles were sorted and screened for eligibility in two stages. The first stage involved title and abstract screening, in which topics had to address rabies using a spatial and/or spatial-temporal approach and met all inclusion criteria. In the next stage, all articles relevant to the topic were screened based on the research objectives, methods, results, and conclusions. Articles that did not meet one or more of the criteria were excluded. The exclusion criteria are described in the PRISMA diagram (Figure 1). Two independent researchers conducted this process to ensure objectivity.
Figure 1: Selection process used in a systematic review of Geospatial analysis applied to rabies epidemiological studies, 2014-2024.
Data Extraction
The data were extracted from selected studies using a custom-designed worksheet. The following basic information was extracted: a) basic information, including title, author, year of publication (years), journal (name, volume, issue, pages, and journal indexing); b) study design, including study objectives, type of study used, research location, and population studied (human and animal rabies epidemiology); c) geospatial methods, including the type of geospatial data used, type of data used for analysis, software, and spatial analysis techniques chosen; and d) main results, including the main findings related to spatial or spatial temporal distribution, low spots and hotspots, level of vulnerability and/or risk of rabies in an area. The data were extracted by two independent researchers. Any discrepancies will be discussed until a consensus is reached, and a third researcher will be involved, if necessary.
Quality Assessment
The methodological quality of each selected article was assessed using a predetermined checklist in the form of a Joanna Briggs Institute (JBI) or Newcastle‒Ottawa Scale (NOS) worksheet [11]. This assessment will help to assess the reliability and validity of the findings reported in previous studies.
Data Collection and Analysis
Two researchers independently conducted this stage. The extracted data were descriptively analyzed to provide an overview of the characteristics of the studies included in this review. This analysis will include the frequency of use of specific geospatial methods, geographical distribution of the studies, and main patterns of findings expressed in the studies, including the title and objectives of the research. Key results from the selected studies were analyzed qualitatively to identify key themes and a narrative synthesis of the application of geospatial analysis in rabies epidemiology. The main focus of the studies included data analysis units, typology of representation, spatial methods used (geospatial methods), and key outcomes of spatial analysis in rabies (contribution to understanding rabies epidemiology and the challenges faced). The results of this systematic review are reported in accordance with the PRISMA guidelines and include a PRISMA flowchart [10] to illustrate the study selection process, tables summarizing study characteristics and results, and an in-depth discussion of the implications of the findings for rabies control practice and future research.
Results
Search Results
The results of the literature search and screening yielded 38 articles that met the inclusion and exclusion criteria from 161 articles. These articles were published between 2014 and 2023. The stepwise screening of articles based on these criteria is presented in the PRISMA chart and PICOS framework (Figure 1).
Study Summary
Initially, 161 articles, published in English, between 20014 and 2024, were selected. After reading the abstracts, 83 articles were excluded because they did not satisfy the inclusion criteria. Only 38 of the other 83 articles were found to meet the review criteria.
We found that some authors cited spatial analysis in the abstract but did not use a spatial method to analyze the data in the geographic information system [12-14]. For example, some articles had incorporated the term “spatial” but used fluorescence micro-optical sectioning tomography (fMOST) as it applies to neurologic [14]. Furthermore, used the logistic regression model to develop an investigation for socio-spatial [12] and to create a new application to dog rabies as a graph-based evidence synthesis approach to detecting outbreak clusters [13]. A comparison of the spatial analysis methods used in the selected articles is given in Table 1.
Table 1: Review of the spatial analysis method used in the selected articles.
| No. | Reference | Spatial Method | Objectives of Spatial Analysis |
| 1 | [7] | Kernel density estimation [15], SaTScan [16]. | To describe spatial and temporal patterns of rabies. To discuss effectiveness of oral rabies vaccination campaigns in targeted oblasts. To identify geographical territories with zero rabies cases among animals. To detect spatial-temporal clusters of rabies cases using space-time permutation model. To compare densities of disease cases on an annual basis. |
| 2 | [8] | Spatial methods include Moran's I, Geary's C [17], and Getis-Ord's G[18]. | To visualize disease data using GIS tools to show spatial distribution. To study relationship between disease incidence and environmental factors geographically. To analyze spatial dynamics to understand spread and develop control strategies. |
| 3 | [9] | Geostatistics estimate and predict [19]. | To determine relationship between temperature, precipitation, and rabies spatial distribution. To identify high-risk areas for rabies outbreaks using geographic information systems. To predict spatial risk and distribution of rabies cases in livestock. |
| 4 | [20] | Moran's global index and Cluster and Outlier Analysis [17]. | To identify clusters of high and low values in rabies cases. To classify governorates based on the number of rabies or PEP cases. |
| 5 | [21] | SaTScan [16], Global Polynomial Interpolation and Contour tool [22]. | To identify high-risk clusters and spatial expansion of human rabies cases. To analyze spatial distribution and clustering of human rabies occurrences. To determine trends in the spread speed of human rabies cases. |
| 6 | [23] | Bayesian hierarchical spatiotemporal model [24] | To understand spatiotemporal variation of rabies. To examine impacts of environmental, economic, and demographic factors on rabies |
| 7 | [25] | Monte Carlo simulations [26]. | To assess spatial association between rabid dogs and urban structures. To determine if rabid dogs are closer to water channels than expected. To analyze spatial clustering of rabid dogs. |
| 8 | [27] | Average Nearest Neighbor (ANN) method [28]. | To determine disease distribution patterns for effective control strategies. To analyze spatial data to identify clustering, dispersion, or randomness. To measure distances between objects to understand distribution patterns. |
| 9 | [29] | Negative binomial regression [30], Z-score analyses [31]. | To assess behavior and status of raccoon variant rabies virus cases. To examine virus in non-raccoon hosts. To evaluate impact of oral rabies vaccine distribution |
| 10 | [32] | Spatial scan statistics [16], Poisson regression modeled variables [33]. | To identify clusters of canine rabies cases. To evaluate the influence of social determinants on canine rabies incidence. To create a risk map for canine rabies based on social factors |
| 11 | [34] | Statistical modeling of cointegration techniques [35]. | To Verify effect of weather components on rabies incidence. To Establish long-run relationship among reported rabies cases, temperature, and precipitation. To Guide local health officials in formulating preventive strategies for rabies control |
| 12 | [36] | Generalized Additive Models [37], Two-dimensional LOESS [38], Permutation tests [39]. | To identify spatial patterns in data for vaccination campaign planning. To Assess clustering of unvaccinated dogs to improve rabies prevention strategies. To Analyze geolocation impact on canine vaccination. To Maintain confidentiality by avoiding public availability of geographic coordinates. |
| 13 | [40] | Moran’s index [17], Chi- squared tests [41]. | To Compare Myotis species identification using morphological keys and genetic identification. Characterize temporal and spatial trends of bat RABV. Assess risk factors for bat RABV infection by circumstances of encounter. |
| 14 | [42] | Used discrete Poisson spatial model [43], Using space-time permutation model [44]. | To Identify clusters of high rabies incidence in specific regions. To Determine areas with significant aggregation of rabies cases in dogs. |
| 15 | [45] | WGS techniques [46]. | To identify genetic relationships of RABV variants, reservoirs, and spatial origin. To generate novel data for future investigations. To monitor virus evolution, transmission, and emergence of relevant genetic mutations |
| 16 | [47] | Phylogeny reconstructed by maximum likelihood and Bayesian methods [48]. Beast and SPREAD software[49]. | To understand evolutionary history and spatial temporal dynamics of rabies virus. To determine evolutionary rates and phylogeographic using Beast and SPREAD software. To illustrate the evolution history and phylogeographic of RABV in badgers. |
| 17 | [50] | Continuous time and space [51]. | To identify patterns in time and space for fox rabies control. To focus on disease dynamics in continuous time and space domains. To highlight potential risk areas and need for effective rabies vaccination |
| 18 | [52] | Spatially explicit individual-based model [53]3 | To investigate how spatiotemporal variation in wildlife host home range movement, implemented as home range size variation, affects the spatial spread, persistence, and incidence of wildlife disease and vaccination effectiveness. |
| 19 | [54] | Used Moran Index [17] and Moran Local Index [55]. | To identify clusters of inadequate post-exposure human rabies procedures spatially. To characterize exposure to the disease and its risk factors geographically. To provide epidemiological evidence for health policy planning and decision-making |
| 20 | [56] | Bayesian Markov Chain Monte Carlo (MCMC) simulation in the BEAST [57]. | To infer viral phylogenetic relationships in space and time. To gain insights into the contribution of host population to viral spread. |
| 21 | [58] | Bayesian regression models [48], Used Integrated Nested Laplace Approximations (INLA) [59]. | To assess impact of accessibility on yearly rate of PEP patients. To generate risk map to identify optimal locations for future centers. To consider travel time to nearest IPC center as primary exposure variable. To investigate spatial and temporal scales for PEP patient predictions. |
| 22 | [60] | Cross-K function [61]. | To analyze spatial clustering between feeding points and dog rabies cases. To investigate the spatial association of stray dog feeding behavior and rabies cases. To simulate the spread of rabies virus among dogs in different subpopulations |
| 23 | [62] | Generalized Additive Models (GAM) [37], SaTScan [16]. | To identify risk factors for rabies. To predict spatial risk areas for rabies spread. To quantify association between monthly rabies occurrences and explainable variables. To Strengthen surveillance system in high-risk areas. |
| 24 | [63] | DOT map cartograms [64]. | To evaluate statistical association between administrative divisions and rabies cases. To use DOT density maps for comparing distribution of rabies cases. |
| 25 | [65] | Dynamic patch occupancy [66]. | To predict spatio-temporal dynamics and spread rates in wildlife diseases. To understand factors driving spread like seasonality, transmission distance, and infection density. To estimate transmission distance, rates of spatial spread, and direction of invasion. |
| 26 | [67] | Logistic regression [68]. | To identify trends in animal bites and rabies incidence spatially. To analyze spatial distribution of animal bites and pets vaccinated. To determine regions with higher incidence of animal bites. To assess spatial patterns of rabies-related human mortality. |
| 27 | [69] | Kernel function used for spatial distribution analysis [70], Multi-criterion Analytical Hierarchy Process technique [71]. | To identify high-risk areas for bovine/human rabies transmitted by bats. To analyze spatial distribution of rabies cases. To assess association with independent variables. To Evaluate environmental and socioeconomic variables for dynamic epidemiological mosaic. |
| 28 | [72] | Smartphone technology directed [73]. | To identify clusters of rabies cases for targeted interventions. To analyze spatial distribution of rabies cases for epidemiological insights. To understand geographic patterns of rabies transmission for control strategies. |
| 29 | [74] | SaTScan [16], Choropleth maps[75], Phylogenetic tree [76]. | To identify spatial clusters of dog rabies cases for analysis. To understand spatial relationships between animal and human rabies cases. To compare positivity rates in different provinces using spatial analysis |
| 30 | [77] | General Method of Moments (GMM) [78], Spatial autocorrelation addressed [79]. | To quantify socio-economic and climate factors in spatial distribution of rabies. To understand spatial heterogeneity and spatial dependence effects in regression models. To analyze the influence of climatic and socioeconomic factors on rabies spread. To compare traditional regression models with aggregation model for better performance. |
| 31 | [80] | Chi- squared tests [41], Moran’s index [55]. | To assess geographic and temporal trends in human and animal rabies cases. To identify provinces with higher bite rates and human rabies cases. To Determine the distribution of human suspect rabies cases throughout the country. |
| 32 | [81] | Kriging method [82], Geostatistics[19]. | To identify geographic clusters of rabies for control strategies. To understand spatial distribution patterns of animal rabies. To highlight landscape determinants of the disease (rural, urban, suburb). |
| 33 | [83] | Descriptive statistics [84]. | To describe human rabies incidence and spatial distribution. To investigate secondary cases and suggest pre-exposure prophylaxis for high-risk populations |
| 34 | [85] | Inverse Distance Weighted Interpolation [86], Getis-Ord's Gi statistic [18]. | To identify risk factors and patterns of human rabies exposure. To analyze spatial distribution of human rabies exposures using GIS tools. To provide insights for cost-effective disease prevention and control measures. |
| 35 | [87] | Quasi-Binomial Regression Model [88]. | To assess current risk of rabies spread. To evaluate efficacy of rabies contingency plans. To analyze influence of dog owner responses to rabies incursions. |
| 36 | [89] | Spatial method [90]. | To clarify epidemiology of rabies. To evaluate factors influencing spread of rabies. To assess effects of rabies control and preventive measures. |
| 37 | [91] | Chi- squared tests [41]. | To identify spatial patterns, relationships, and trends in dog biting incidents. To determine the association between dog bites and neighborhood characteristics. To analyze the spatial distribution of dog bites based on socioeconomic factors. |
| 38 | [92] | Kernel density estimation [15]. | To visualize bat collection points and distribution patterns. To estimate density contribution of each point in the analysis. To identify statistically important areas for bat rabies surveillance. To analyze seasonal variation in bat removal requests and testing samples. To assess the spatial relationship between warmer months and bat activity. |
Year of Studies and Publication
Variations were reported in the period studied. Most published studies involved an analysis of data covering one year or more. Generally, the articles used data that had been collected one to six years before publication. About 53.8 % of the studies published used data collected for 1 to 5 years and 17.9 % was used the data collected for 6 to 10 years. Furthermore, more than 82.1 % of the studies were published just less than five years after the event occurred, and only 2.6 % of the studies used data collected more than 30 years before publication.
Not many papers using spatial analysis [25, 27, 47, 50, 56, 65, 69, 89] were published between 2014 and 2017, but since 2018 the number of papers based on geospatial studies has increased. Approximately 60 % of the relevant papers were published after 2018 [7-9, 14, 20, 21, 23, 27, 29, 32, 34, 36, 40, 42, 45, 52, 54, 58, 60, 62, 63, 67, 72, 74, 77, 80, 81, 83, 85, 87, 91, 92] (Table 2).
Most of the studies were undertaken by USA and Brazilian or American investigators. These countries are responsible for 55% of all the studies developed, followed by China, Tunisia, Thailand and United Kingdom (UK). However, it is noteworthy that, in the case of USA and Brazil, the studies were carried out with each database for rabies in their institutions. In the United Kingdom (UK), on the other hand, the studies were undertaken using other countries’ databases [56], [72].
The articles were published in the various journals listed in Table 2, most of them in just five journals: PLoS Neglected Tropical Diseases, PLOS ONE, Journal of Geospatial Health, Frontiers in Veterinary Science, and Tropical Medicine and Infectious Disease. Furthermore, mostly the reputation of articles in Scopus and Web of Science is in the form of Quartile (Q1), followed by a Quartile 2 (Q2) and a Quartile 3 (Q3). These journals published 49% of the articles that used spatial analysis methods in the investigation of rabies transmission.
Table 2: Total articles by year and periodical.
| No. | Year | Number of Article | Periodical | Quartile |
| 1 | 2014 | 1 | Emerging Infectious Diseases | https://wwwnc.cdc.gov/eid/about Scopus Quartile 1 (Q1) |
| 2 | 2015 | 1 | Geospatial Health | https://www.geospatialhealth.net/ Scopus Quartile 3 (Q3) |
| 3 | 2016 | 3 | PLOS Neglected Tropical | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) |
| PLOS ONE journal | https://journals.plos.org/plosone/ Scopus Quartile 1 (Q1) | |||
| Molecular Sciences MDPI | https://www.mdpi.com/journal/ijms Scopus Quartile 1 (Q1) | |||
| 4 | 2017 | 2 | PLOS Neglected Tropical Diseases, | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) |
| Tropical Medicine and Infectious Disease. | https://www.mdpi.com/journal/tropicalmed Scopus Quartile 2 (Q2) | |||
| 5 | 2018 | 7 | PeerJ, | https://peerj.com/ Scopus Quartile 2 (Q2) |
| PLOS ONE, PLOS ONE, | https://journals.plos.org/plosone/ Scopus Quartile 1 (Q1) | |||
| BMC Infectious Diseases, | https://bmcinfectdis.biomedcentral.com/ Scopus Quartile 1 (Q1) | |||
| Journal Epidemiologia e servicos de saude, | https://www.scielo.br/j/ress/ Scopus Quartile 2 (Q2) | |||
| Epidemiology and Infection journal, | https://www.cambridge.org/core/journals/epidemiology-and-infection Scopus Quartile 2 (Q2) | |||
| BMC Veterinary Research | https://bmcvetres.biomedcentral.com/ Scopus Quartile 1 (Q1) | |||
| 6 | 2019 | 5 | Frontiers in Veterinary Science, | https://www.frontiersin.org/journals/veterinary-science Scopus Quartile 1 (Q1) |
| Geospatial Health, | https://www.geospatialhealth.net/ Scopus Quartile 3 (Q3) | |||
| PLOS Neglected Tropical Diseases, | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| Journal of the Brazilian Society of Tropical Medicine, | https://www.scielo.br/j/rsbmt/ Scopus Quartile 3 (Q3) | |||
| Ethiopian Veterinary Journal. | https://www.ajol.info/index.php/evj Scopus Quartile 1 (Q1) | |||
| 7 | 2020 | 5 | PLOS ONE, | https://journals.plos.org/plosone/ Scopus Quartile 1 (Q1) |
| Preventive Veterinary Medicine, | https://www.sciencedirect.com/journal/preventive-veterinary-medicine Scopus Quartile 1 (Q1) | |||
| Journal of Animal Ecology, | https://www.scirp.org/journal/oje/?utm Scopus Quartile 1 (Q1) | |||
| Frontiers in Veterinary Science, | https://www.frontiersin.org/journals/veterinary-science Scopus Quartile 1 (Q1) | |||
| PLOS Neglected Tropical Diseases | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| 8 | 2021 | 7 | Veterinary Medicine and Science, | https://onlinelibrary.wiley.com/journal/20531095 Scopus Quartile 2 (Q2) |
| Veterinary World, | https://www.veterinaryworld.org/ Scopus Quartile 2 (Q2) | |||
| viruses MDPI Journal, | https://www.mdpi.com/journal/viruses Scopus Quartile 1 (Q1) | |||
| PLOS Neglected Tropical Diseases | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| Tropical Medicine Infection Disease | https://www.mdpi.com/journal/tropicalmed Scopus Quartile 2 (Q2) | |||
| PLOS Neglected Tropical Diseases, | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| Heliyon Journal | https://www.cell.com/heliyon/home Scopus Quartile 1 (Q1) | |||
| 9 | 2022 | 6 | Journal of Geospatial Health, | https://www.geospatialhealth.net/ Scopus Quartile 3 (Q3) |
| Environmental Research and Public Health, | https://www.mdpi.com/journal/ijerph Scopus Quartile 2 (Q2) | |||
| Clin Transl Med, | https://onlinelibrary.wiley.com/journal/20011326 Scopus Quartile 1 (Q1) | |||
| PLOS Neglected Tropical Diseases, | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| NATURE COMMUNICATIONS, | https://www.nature.com/ncomms/ Scopus Quartile 1 (Q1) | |||
| PLOS Neglected Tropical Diseases. | https://journals.plos.org/plosntds/ Scopus Quartile 1 (Q1) | |||
| 10 | 2023 | 2 | Tropical Medicine and Infectious Disease (MDP), | https://www.mdpi.com/journal/tropicalmed Scopus Quartile 2 (Q2) |
| International Journal of Infectious Diseases | https://www.scirp.org/journal/aid/?utm_campaign Scopus Quartile 1 (Q1) |
Human and Animal Rabies Epidemiological Information
Nine of the studies included in this review applied spatial methods to analyze epidemiological information about human and animal rabies [9, 20, 58, 63, 69, 74, 80, 87, 91],and nine articles just analyzed epidemiological information about human rabies [8, 21, 23, 54, 72, 77, 83, 85, 89], and 21 articles focused solely on the epidemiology of animal rabies [7, 14, 25, 27, 29, 32, 34, 36, 40, 42, 45, 47, 50, 52, 56, 60, 62, 65, 67, 81, 92].
In terms of the geometric or shape representation of data, the studies primarily used polygons and point data. The polygons were used to represent administrative frontiers, such as neighborhoods, districts or other administrative frontiers, the points were used to represent cases of rabies, households, animals (dog, cat, Bats etc.) traps.
There is no predominant type related to the topology utilized because it was common to use more than one type in the articles. For example, often data are collected at household level, but for analysis purposes they are aggregated into areas.
Spatial Units
In the articles selected, 12 different primary units of analysis were identified. The most-used primary unit of analysis was the more than one unit of spatial analysis; for examples counties, cities, provinces, district, human and animal rabies cases, municipalities, administrative town, city, river, lake and department, and village which were applied in fifteen articles, or around 38% of the published studies [9, 21, 72, 80, 81, 89, 92, 27, 32, 36, 40, 42, 58, 62, 67]. The human and rabies cases as primary unit was used in seven studies [14, 50, 52, 60, 63, 65, 77], and Municipalities were used in four studies [54, 69, 74, 83]. Two studies [23, 7] used the province as the unit of analysis. Other studies used the region as analysis unit [56, 47]. In two studies, City were used [20, 34]. State was applied in two studies [87, 45], administrative districts were used in one [85], and the Counties unit was used in one [29], the global units was used in one [8], and neighborhood unit was used in one study [91]. Furthermore, the line (water channel) unit was used in one study [25].
Methods of Spatial Analysis Applied in Rabies Studies
Twenty-eight different spatial methods used to analyze rabies data were found in the articles. However, some were more common than others. The methods used in selected papers are listed according to the topology of the data used (Table 1).
Spatial Analysis of Points
In the analysis of point data, the method used most frequently, in 5 papers [32, 42, 62, 72, 74], was Spatial scan statistics (SaTScan) [16]. The Moran's index statistic [55] was used in two papers [8], [77]. The Kernel density analysis [15] was applied in a separate study [92] and in another study [7], the standard distances, space-time permutation model were applied. Two papers [23, 58] used only the Bayesian models [48]. The Getis-Ord Gi statistic [18], was used in two studies [8, 85].
The ANN (average nearest neighbor) [28] was applied in one paper [27]. The Discrete Poisson spatial model (DPS) [43] was applied in one study [42] and the Cross-K function [61] was applied in two papers [36, 50]. The Kriging method [82], and Geostatistics [19] were applied in two papers [9, 81]. The Monte Carlo simulations [26] and the L function [93] were used in one paper [25]. The Standard distances [94], space-time permutation model [44] were applied in one paper [7]. In one paper [8], Geary's Contiguity Ratio [95] was used. The analytical Hierarchy Process (AHP) technique [71] was applied in one [69], the R for accuracy test [96] was used in another [52] and, DOT map cartograms reshape [64] was applied in one study [63]. Furthermore, the fMOST and scRNA-seq techniques reveal 3D rabies virus (RABV) distribution in the brain [97] was used in one study [14]. Finally, the descriptive statistic model [84] was applied in one study [83].
Spatial analysis of area data
SaTScan (spatial scan statistics) [16] was the method used most often in analyzing polygon data [32], [42, 72, 74]. Another method commonly used was the Global Moran Index [55] which was applied in three studies [20, 54, 77]. The Cross-K function [61] were applied in two studies [50, 60]. Furthermore, the Generalized Additive Models (GAM) [78] was applied in two studies [36, 62]. The Monte Carlo method [26] was applied in one study [25], the average nearest neighbor (ANN) [28] method in another [27], the Kriging method [82] was applied in one study [81], the Bayesian regression models [48] was applied in one study [58] and the Getis-Ord Gi statistic [18] in another [91].
Software Programs Used for Spatial Analysis of Rabies Cases
Some articles did not report which software had been used to perform the spatial analysis of data. Further, in some cases, the method of spatial analysis was not referenced; instead, the focus was on the set of operations utilized. For example, it was clear in every article that different software had been used; in some cases, one software program was used to create the geographical coordinates (latitude and longitude), and another specifically to perform the spatial analysis. The software programs used in the selected articles are given in (Table 3). The most used were ArcGIS, R software, GeoDa, TerraView, Moran's index and MapInfo. Several other software programs - for example, Satscan, Terrasee, BioEdit software, ClustalX version 1.8. MegAlign software version 5. MEGA version 5, R and the stpp-package Cellular automata (CA) and other customized ones - were used, but not as often.
Table 3: List the software used each year.
| No. | Year | Software | Number of Studies |
| 1 | 2014 | BioEdit software, ClustalX version 1.8. | 1 |
| MegAlign software version 5. | 1 | ||
| MEGA version 5 | 1 | ||
| 2 | 2015 | R and the stpp-package | 1 |
| Cellular automata (CA) | 1 | ||
| 3 | 2016 | Kernel function | 1 |
| TerraView 4.2.2 | 1 | ||
| ArcGis 10.0 | 1 | ||
| Beast and SPREAD | 1 | ||
| Maximum likelihood and Bayesian | 1 | ||
| 4 | 2017 | R software | 1 |
| Geographic Information System (GIS) | 1 | ||
| ArcGIS 10.3 | 1 | ||
| Dynamic patch-occupancy model | 1 | ||
| 5 | 2018 | R (version 3.4.2) with glmmADMB package | 1 |
| The glmmADMB package | 1 | ||
| SaTScan(tm) v8.0 software | 1 | ||
| SAS 9.4 and Microsoft Excel 2013 | 1 | ||
| GMM adopted for unbiased estimation with spatial autocorrelation. | 1 | ||
| Moran's I statistic | 1 | ||
| Tab Win 32, Epi Info 7.1, and Microsoft Excel 2010. | 1 | ||
| Individual-based model on a 1-by-1 km grid. | 1 | ||
| Spatially explicit transmission model developed at 1 km2 grid scale | 1 | ||
| Minitab software for descriptive statistics and linear regression analysis. | 1 | ||
| Kernel density estimation. | 1 | ||
| 6 | 2019 | R package malaria Atlas | 1 |
| Rapid Extractor of Climatological Information III (ERIC III). | 1 | ||
| Geographic Information Systems (ArcGIS) software | 4 | ||
| TerraView 4.2.2 | 1 | ||
| Moran's index. | 1 | ||
| 7 | 2020 | Spatial analysis conducted using Moran's I statistic for geographic trends | 1 |
| Cross-K function. | 1 | ||
| Metapopulation analysis. | 1 | ||
| SEIR model. | 1 | ||
| Spatially explicit individual-based model. | 1 | ||
| Sensitivity analyses conducted | 1 | ||
| SaTScan software utilized for spatial and spatio-temporal analysis. | 1 | ||
| No information | 1 | ||
| Moran's global index and spatial autocorrelation by Cluster and Outlier Analysis | 1 | ||
| 8 | 2021 | Package "spdep" of R software for spatial analysis | 1 |
| ArcGIS was used for spatial analysis | 4 | ||
| SaTScan. | 1 | ||
| Ordinary Kriging regression. | 1 | ||
| Kriging method applied to estimate missed data in reporting process. | 1 | ||
| Generalized Additive Models (GAMs) were used for spatial analysis | 1 | ||
| 9 | 2022 | Dynamical models and phylogenetic analysis | 1 |
| ArcGIS | 2 | ||
| SaTScan version 9.3 | 1 | ||
| fMOST and single-cell RNA sequencing techniques | 1 | ||
| Spatio-temporal Bayesian regression models | 1 | ||
| Bayesian statistics. | 1 | ||
| R code | 1 | ||
| SaTScan. | 2 | ||
| 10 | 2023 | ArcGIS software | 2 |
| Stata version 17 | 1 |
Discussion
Despite place being a fundamental component of epidemiological investigations, the small number of papers found may indicate that the use of spatial analysis in studies of rabies is still uncommon. Among the possible reasons that may hinder the application of spatial analysis in the data analysis of rabies is the lack of health information systems that produce georeferenced human and animal epidemiological information, that is, the appropriate scales of analysis.
As of 2018, there was a significant increase in the number of articles that addressed the application of spatial analysis in studies of rabies. 60% of the published articles were found from that year on, perhaps due to the increased severity of the rabies epidemics the world observed during this period. The increase could also be a result of the need to develop new approaches to rabies research, to better understand the dynamics of the disease’s transmission, and to formulate strategies to minimize its effects.
Among the works selected, there was a significant time interval between the occurrence of events (rabies cases), or even between the execution of virological and human and animal (zoonotic) surveys, and the publication of the results of spatial analyses. This time lag might have been due to the natural flow of research, but it might also reflect the complexity and difficulties involved in conducting spatial analyses in countries where rabies is endemic.
The existence of such a long-time interval between the collection of humans and animals’ epidemiological information and the analysis and dissemination of results can lead to some bias against the early detection of epidemics [98]. Therefore, there is a reduction in the ability to identify the surveillance sites that require public health action.
Although most studies of rabies using spatial analysis were based on data from countries where the disease is endemic, the studies themselves were conducted elsewhere. The exception is the USA, which has developed most of the spatial analyses, according to articles published in the countries themselves. Brazil and China figured most among the countries studied, with 26% of the articles, followed by Colombia, Peru, and Tunisia, which are often targeted for research because they highlight geographical disparities in research focus and capability, but countries with geographical disparities in rabies, such as Timor-Leste [99], Indonesia [100], and others that are highly zoonotic areas, have not been widely reported. Therefore, research focus and capability in geospatial analysis are necessary in other areas with widely reported zoonotic diseases, particularly rabies cases.
Furthermore, more than one unit of spatial analysis has been the most commonly used as a spatial unit, followed by human and rabies cases as primary units and municipalities. This could be due to the actual characteristics of the epidemiological studies of human and animal rabies, which often focus on the areas surrounding (the counties, cities, provinces, districts, human and animal rabies cases, municipalities, administrative towns, cities, rivers, lakes, departments, villages, etc.) because of the ecological characteristics associated with the spread of animals (dogs, mice, bats, etc.) [3, 6, 74], that were the transmitter agents of rabies disease.
Twenty-eight different methods of spatial analysis were found; however, only two of them were used frequently. In analyzing the point data, the methods most used have been the SaTScan (spatial-time scan). These methods are essentially imposing circles of different sizes (from zero up to a defined proportion of the population size) on the geographic area and for each circle, computes a likelihood ratio statistic based on the number of observed and expected cases [16].
For the analysis of the polygon data or areas, the most widely used indicator is SaTScan (spatial-time scan) too, and Moran’s global statistic is the commonest way of measuring the degree of spatial autocorrelation in area data [101], perhaps because of the ease of application and interpretation of results, greater availability of free ArcGIS, R software, GeoDa, TerraView, Moran's index and MapInfo, or the lack of health information on more greatly detailed scales.
The other twenty-five methods of analysis were each used by only one or two articles, and always as a complement to traditional methods of statistical analysis.
Some of the methods might have been more used due to their greater popularization, because of ease of access, as generally they are implemented in commercial software of great diffusion capacity with easy-to-handle friendly interfaces, as also in those of the public domain, with a larger number of courses and tutorials which assist the user in their use. On the other hand, although some of the methods least used may be available, both in software of the public domain as also in commercial software, they have as yet been little disseminated and present little didactic material, often having relatively unfriendly interfaces which make their manipulation on the part of users who have little familiarity with special data, difficult. Although they are commercial software, ArcView/ArcGIS and MapInfo are among the software programs most used, probably because they present more friendly interfaces than do the free software programs and count on a wide range of programs for the dissemination of and training in their use. On the other hand, although they are free (software) programs, GeoDa and TeraView are also frequently used, probably because they make the methods commonly used in spatial analyses in public health available free, together with didactic material, and are widely represented at technical and scientific events [102-104].
Despite these considerations, it is pertinent to point out that regardless of the degree of sophistication of the method used, the results shown in the papers pointed to the great utility of spatial analysis for the understanding the epidemiology of human and animal rabies cases on different continents and in different geographical areas. Furthermore, results have shown that the methods such as SaTScan (spatial-time scan), and Moran’s global, can quickly produce efficient information regarding the location of clusters of rabies cases and of hotspot areas of transmission [16, 44, 101]. Such information can be a powerful tool for monitoring rabies transmission at the local level.
Finally, despite the development of the methods of spatial analysis applied in epidemiological studies, they are rarely used in studies of rabies. However, the identification of spatial patterns in most of the articles discussed above confirms the usefulness of the application of these techniques and the need for development and application of advanced spatial analysis beyond the limits of visualization.
Some spatial analysis methods should be used in conjunction with conventional methods as, for example, in the control diagrams currently used by public health programs to identify rabies risk [105]. The use of these methods to advance scientific knowledge on the dynamics of rabies transmission and its spatial diffusion could certainly be incorporated into current surveillance strategies and may contribute to reducing social costs, by incorporating both the individual and contextual variables associated with rabies transmission.
Conclusion
Despite its potential, spatial analysis remains underutilized in rabies studies, largely because of the lack of georeferenced epidemiological data for humans and animals. Since 2018, there has been a significant increase in the number of studies that have used spatial analysis in rabies research. This increase is likely driven by the severity of rabies epidemics and the need for innovative research approaches to understand and mitigate disease transmission. These findings suggest that there is a significant delay between rabies incidence, data collection, and the subsequent publication of spatial analysis results. This delay may hinder the early detection of epidemics and timely public health interventions.
Most spatial analysis studies on rabies are conducted in countries where the disease is endemic; however, the research itself is often conducted in other countries. The United States is an exception, as it has domestically conducted most of its spatial analyses. Countries such as Brazil and China are often targeted for research because they highlight geographical disparities in research focus and capability, but countries with geographical disparities in rabies, such as Timor-Leste, Indonesia, and others that are highly zoonotic areas, have not been widely reported.
Although 28 different spatial analysis methods have been identified, only a few have been used in countries where the results are widely accepted. Methods such as the SaTScan and Moran indices are popular because of their availability in commercial (ArcGIS and MapInfo) and free (GeoDa and TerraView) software with supporting didactic materials. In the future, advanced spatial analysis methods should complement conventional epidemiological tools such as control charts used in public health programs.
Integrating these methods can improve our understanding of rabies transmission dynamics and provide more effective surveillance and intervention strategies. There is a clear and urgent need to develop and apply spatial analysis techniques for rabies research and eradication in the region. By identifying spatial patterns and incorporating individual and contextual variables, these methods can significantly contribute to reducing the social and economic impacts of rabies, including specific geographic and health disparities.
References
- G M, Kavitha, Chethana Chandrahasa, Akshay Dangari, et al. (2023). Comprehensive Update on Rabies: A Neglected Zoonotic Disease of Public Health Concern. Progress In Microbes & Molecular Biology. 6(1):1-21.
Publisher | Google Scholor - O. Article et al., (2021). Narra J Cerebellum, 2(July):1-11.
Publisher | Google Scholor - Fooks, A., Cliquet, F., Finke, S. et al. (2017). Rabies. Nat Rev Dis Primers, 3:17091.
Publisher | Google Scholor - Jane Ling MY, Halim AFNA, Ahmad D, et al, (2023). Rabies in Southeast Asia: A Systematic Review of its Incidence, Risk Factors and Mortality, BMJ Open, 13:e066587.
Publisher | Google Scholor - Gan H, Hou X, Wang Y, Xu G, Huang Z, et al. (2023). Global Burden of Rabies In 204 Countries and Territories, From 1990 To 2019: Results from The Global Burden of Disease Study 2019. Int J Infect Dis. 126:136-144.
Publisher | Google Scholor - Singh, R., Singh, K. P., Cherian, S., Saminathan, M., Kapoor, S., et al. (2017). Rabies - Epidemiology, Pathogenesis, Public Health Concerns and Advances in Diagnosis and Control: A Comprehensive Review. Veterinary Quarterly, 37(1):212-251.
Publisher | Google Scholor - Polupan I, Bezymennyi M, Gibaliuk Y, Drozhzhe Z, Rudoi O, et al. (2019). An Analysis of Rabies Incidence and Its Geographic Spread in the Buffer Area Among Orally Vaccinated Wildlife in Ukraine from 2012 to 2016. Front Vet Sci. 6:290.
Publisher | Google Scholor - Chen, S. (2022). Spatial And Temporal Dynamic Analysis of Rabies: A Review of Current Methodologies. Geospatial Health, 17(2).
Publisher | Google Scholor - Bárcenas-Reyes I, Nieves-Martínez DP, Cuador-Gil JQ, Loza-Rubio E, González-Ruíz S, et al. (2019). Spatiotemporal Analysis of Rabies in Cattle in Central Mexico. Geospat Health. 14(2).
Publisher | Google Scholor - M. J. Page et al., (2021). The Prisma 2020 Statement: An Updated Guideline for Reporting Systematic Reviews, BMJ, 372:71.
Publisher | Google Scholor - Checklist for Prevalence Studies.
Publisher | Google Scholor - Sánchez-Soriano, C., Gibson, A.D., Gamble, L. et al. (2019). Development of A High Number, High Coverage Dog Rabies Vaccination Programme in Sri Lanka. BMC Infect Dis. 19:977.
Publisher | Google Scholor - Cori A, Nouvellet P, Garske T, Bourhy H, Nakouné E, et al. (2018). A Graph-Based Evidence Synthesis Approach to Detecting Outbreak Clusters: An Application to Dog Rabies. PLoS Comput Biol. 14(12):e1006554.
Publisher | Google Scholor - Zhang Y, Xing X, Long B, Cao Y, Hu S, et al. (2022). A Spatial and Cellular Distribution of Rabies Virus Infection in The Mouse Brain Revealed by FMOST and Single-Cell RNA Sequencing. Clin Transl Med. 12(1):e700.
Publisher | Google Scholor - Y. Chen, (2018). Lec6_hist_KDE, 1-11.
Publisher | Google Scholor - Daan Vink, (2018). Local cluster detection using spatial scan statistics.
Publisher | Google Scholor - Anselin, Luc. (1995). Local Indicators of Spatial Association-ISA. Geographical Analysis. 27:93-115.
Publisher | Google Scholor - T. Monzur, (2015). Local G statistics Calculation.
Publisher | Google Scholor - Introduction to Geostatistics Ardossy.
Publisher | Google Scholor - Kalthoum S, Guesmi K, Gharbi R, et al. (2021). Temporal and Spatial Distributions of Animal and Human Rabies Cases During 2012 And 2018, In Tunisia. Vet Med Sci. 7:686-696.
Publisher | Google Scholor - Yue Y, Chen Q, Mu D, Li Y, Yin W. (2022). A Descriptive Analysis of Human Rabies in Mainland China, 2005-2020. Int J Environ Res Public Health. 20(1):380.
Publisher | Google Scholor - T. Methods, T. Geospatial, W. Flow, (2012). Characteristics of Interpolation Methods.
Publisher | Google Scholor - Hangyu Li, Yanjiao Li, Yue Chen, Bo Chen, Qing Su, et al. (2023). Mapping Rabies Distribution in China: A Geospatial Analysis of National Surveillance Data, International Journal of Infectious Diseases, 131:140-146.
Publisher | Google Scholor - Wikle, C. K., Milliff, R. F., Nychka, D., Berliner, L. M. (2001). Spatiotemporal Hierarchical Bayesian Modeling Tropical Ocean Surface Winds. Journal of the American Statistical Association, 96(454):382-397.
Publisher | Google Scholor - Castillo-Neyra R, Zegarra E, Monroy Y, Bernedo RF, Cornejo-Rosello I, et al. (2017). Spatial Association of Canine Rabies Outbreak and Ecological Urban Corridors, Arequipa, Peru. Tropical Medicine and Infectious Disease. 2(3):38.
Publisher | Google Scholor - M. Fippel, (2016). Basics of monte carlo simulations, Monte Carlo Tech. Radiat. Ther., 17-28.
Publisher | Google Scholor - Melyantono SE, Susetya H, Widayani P, Tenaya IWM, Hartawan DHW. (2021). The Rabies Distribution Pattern on Dogs Using Average Nearest Neighbor Analysis Approach in The Karangasem District, Bali, Indonesia, in 2019. Vet World. 14(3):614-624.
Publisher | Google Scholor - George H. Chen, Devavrat Shah. (2018). Explaining the Success of Nearest Neighbor Methods in Prediction, Foundations and Trends® in Machine Learning, 10( 5-6):337-588.
Publisher | Google Scholor - Plants KB, Wen S, Wimsatt J, Knox S. (2018). Longitudinal Analysis of Raccoon Rabies In West Virginia, 2000-2015: A Preliminary Investigation. PeerJ. 6:e4574.
Publisher | Google Scholor - Hilbe JM. (2011). Negative Binomial Regression. 2nd ed. Cambridge University Press.
Publisher | Google Scholor - Curtis AE, Smith TA, Ziganshin BA, Elefteriades JA. (2016). The Mystery of the Z-Score. Aorta (Stamford). 4(4):124-130.
Publisher | Google Scholor - Arias-Orozco P, Bástida-González F, Cruz L, Villatoro J, Espinoza E, et al. (2018). Spatiotemporal Analysis of Canine Rabies In El Salvador: Violence and Poverty as Social Factors of Canine Rabies. PLoS One. 13(8):e0201305.
Publisher | Google Scholor - Chapter 15 Poisson Regression Models, Regression Analysis, 1-3.
Publisher | Google Scholor - Lachica ZPT, Peralta JM, Diamante EO, Murao LAE, Mata MAE, et al. (2020). A Cointegration Analysis of Rabies Cases and Weather Components in Davao City, Philippines from 2006 to 2017. PLoS ONE. 15(8):e0236278.
Publisher | Google Scholor - J. D. Hamilton, (2020). Cointegration, Time Ser. Anal., 1:571-629.
Publisher | Google Scholor - Castillo-Neyra R, Toledo AM, Arevalo-Nieto C, MacDonald H, De la Puente-León M, et al. (2019) Socio-Spatial Heterogeneity in Participation in Mass Dog Rabies Vaccination Campaigns, Arequipa, Peru. PLoS Negl Trop Dis. 13(8):e0007600.
Publisher | Google Scholor - Davis ML, Woodard DA, Pesto M, Toya RE. (1977). Attempts to Purify Hemopoietic Stem Cell Enrichment in Bone Marrow by Use of Glass Wool Filtration. Exp Hematol. 5(4):310-318.
Publisher | Google Scholor - I. Gijbels, I. Prosdocimi, (2010). Loess, Wiley Interdiscip. Rev. Comput. Stat., 2(5):590-599.
Publisher | Google Scholor - N. Haas, (2017). The Two-Sample Problem.
Publisher | Google Scholor - Bonwitt J, Oltean H, Lang M, Kelly RM, Goldoft M. (2018). Bat Rabies in Washington State: Temporal-Spatial Trends and Risk Factors for Zoonotic Transmission (2000-2017). PLoS One. 13(10):e0205069.
Publisher | Google Scholor - D. Mindrila and P. Balentyne, (2013). The Chi -Square Test: Analyzing, Basic Pract. Stat., 23.
Publisher | Google Scholor - Zied Bouslama, Jaber A. Belkhiria, Imed Turki, Habib Kharmachi, (2020). Spatio-Temporal Rvolution of Canine Rabies in Tunisia, 2011–2016, Preventive Veterinary Medicine, 185:105195.
Publisher | Google Scholor - Durán Pacheco, Gonzalo. (2006). Discrete Probability Models to Assess Spatial Distribution Patterns in Natural Populations and An Algorithm for Likelihood Ratio Goodness of Fit Test. Acta Nova, 3(3):543-563.
Publisher | Google Scholor - Kulldorff M, Heffernan R, Hartman J, Assunção R, Mostashari F. (2005). A Space-Time Permutation Scan Statistic for Disease Outbreak Detection. PLoS Med. 2(3):e59.
Publisher | Google Scholor - Hyeon J-Y, Risatti GR, Helal ZH, McGinnis H, Sims M, et al. (2021). Whole Genome Sequencing and Phylogenetic Analysis of Rabies Viruses from Bats in Connecticut, USA, 2018–2019. Viruses. 13(12):2500.
Publisher | Google Scholor - Wyres KL, Conway TC, Garg S, Queiroz C, Reumann M, et al. (2014). WGS Analysis and Interpretation in Clinical and Public Health Microbiology Laboratories: What Are the Requirements and How Do Existing Tools Compare? Pathogens. 3(2):437-458.
Publisher | Google Scholor - Lin Y-C, Chu P-Y, Chang M-Y, Hsiao K-L, Lin J-H, et al. (2016). Spatial Temporal Dynamics and Molecular Evolution of Re-Emerging Rabies Virus in Taiwan. International Journal of Molecular Sciences. 17(3):392.
Publisher | Google Scholor - J. Menčík, (2016). Bayesian Methods, Concise Reliability for Engineers.
Publisher | Google Scholor - Filip Bielejec, Andrew Rambaut, Marc A. Suchard, Philippe Lemey, (2011). SPREAD: Spatial Phylogenetic Reconstruction of Evolutionary Dynamics, Bioinformatics, 27(20):2910-2912.
Publisher | Google Scholor - Eckardt M, Freuling C, Müller T, Selhorst T. (2015). Spatio-Temporal Analysis of Fox Rabies Cases in Germany 2005-2006. Geospat Health. 10(1):313.
Publisher | Google Scholor - Kenji Doya. (2000). Reinforcement Learning in Continuous Time and Space. Neural Comput. 12(1):219-245.
Publisher | Google Scholor - McClure KM, Gilbert AT, Chipman RB, Rees EE, Pepin KM. (2020). Variation in Host Home Range Size Decreases Rabies Vaccination Effectiveness by Increasing the Spatial Spread of Rabies Virus. J Anim Ecol. 89:1375-1386.
Publisher | Google Scholor - Berec, Luděk. (2002). Techniques of Spatially Explicit Individual-Based Models: Construction, Simulation, and Mean-Field Analysis. Ecological Modelling. 150:55-81.
Publisher | Google Scholor - Cavalcante KKS, Florêncio CMGD, Moreno JO, Correia FGS, Alencar CH. (2019). Post-Exposure Human Rabies Prophylaxis: Spatial Patterns of Inadequate Procedures in Ceará - Brazil, 2007 to 2015. Rev Soc Bras Med Trop. 53:e20190247.
Publisher | Google Scholor - Solymosi Reka, Medina Juanjo. (2022). Global and Local Spatial Autocorrelation, Crime Mapp.
Publisher | Google Scholor - Hanke D, Freuling CM, Fischer S, Hueffer K, Hundertmark K, et al. (2016) Spatio-Temporal Analysis of The Genetic Diversity of Arctic Rabies Viruses and Their Reservoir Hosts in Greenland. PLoS Negl Trop Dis. 10(7):e0004779.
Publisher | Google Scholor - Drummond, Alexei; Suchard, M.A.; Xie, D.; Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7, Mol. Biol. Evol., 22:1185-1192.
Publisher | Google Scholor - Baron JN, Chevalier V, Ly S, Duong V, Dussart P, et al. (2022) Accessibility to Rabies Centers and Human Rabies Post-Exposure Prophylaxis Rates in Cambodia: A Bayesian Spatio-Temporal Analysis to Identify Optimal Locations for Future Centers. PLoS Negl Trop Dis. 16(6): e0010494.
Publisher | Google Scholor - Martino, Sara; Riebler, Andrea. (2019). Integrated Nested Laplace Approximations (INLA).
Publisher | Google Scholor - Komol P, Sommanosak S, Jaroensrisuwat P, Wiratsudakul A, Leelahapongsathon K. (2020). The Spread of Rabies Among Dogs in Pranburi District, Thailand: A Metapopulation Modeling Approach. Front. Vet. Sci. 7:570504.
Publisher | Google Scholor - Tao, R., Thill, J. C. (2019). Flow Cross K-Function: A Bivariate Flow Analytical Method. International Journal of Geographical Information Science, 33(10):2055-2071.
Publisher | Google Scholor - Thanapongtharm W, Suwanpakdee S, Chumkaeo A, Gilbert M, Wiratsudakul A. (2021). Current Characteristics of Animal Rabies Cases in Thailand and Relevant Risk Factors Identified by A Spatial Modeling Approach. PLoS Negl Trop Dis. 15(12):e0009980.
Publisher | Google Scholor - Meske M, Fanelli A, Rocha F, Awada L, Soto PC, et al. (2021). Evolution of Rabies in South America and Inter-Species Dynamics (2009-2018). Tropical Medicine and Infectious Disease. 6(2):98.
Publisher | Google Scholor - Soetens L, Hahné S, Wallinga J. (2017). Dot Map Cartograms for Detection of Infectious Disease Outbreaks: An Application to Q Fever, The Netherlands and Pertussis, Germany. Euro Surveill. 22(26):30562.
Publisher | Google Scholor - Pepin KM, Davis AJ, Streicker DG, Fischer JW, VerCauteren KC, et al. (2017). Predicting Spatial Spread of Rabies in Skunk Populations Using Surveillance Data Reported by The Public. PLoS Negl Trop Dis. 11(7):e0005822.
Publisher | Google Scholor - S. Occupancy et al., (2003). Lecture 09 - Patch Occupancy and Patch Dynamics Patch Occupancy: WILD 7970 - Analysis of Wildlife Populations, Sites J. 20Th Century Contemp. French Stud.
Publisher | Google Scholor - Monje F, Kadobera D, Ndumu DB, Bulage L, Ario AR. (2021). Trends and Spatial Distribution of Animal Bites and Vaccination Status Among Victims and The Animal Population, Uganda: A Veterinary Surveillance System Analysis, 2013–2017. PLoS Negl Trop Dis. 15(4): e0007944.
Publisher | Google Scholor - D. Jurafsky, J. Martin, (2012). Logistic Regression, Speech Lang. Process., 404(4):731-735.
Publisher | Google Scholor - de Andrade FAG, Gomes MN, Uieda W, Begot AL, Ramos OdS, et al. (2016). Geographical Analysis for Detecting High-Risk Areas for Bovine/Human Rabies Transmitted by the Common Hematophagous Bat in the Amazon Region, Brazil. PLoS ONE. 11(7):e0157332.
Publisher | Google Scholor - L. B. Engelhardt, (2013). Kernels and Kernel Methods (10/09/13) Kernel Functions, 1-8.
Publisher | Google Scholor - Siekelova, Anna; Podhorska, Ivana; Imppola, Jorma. (2021). Analytic Hierarchy Process in Multiple–Criteria Decision–Making: A Model Example. SHS Web of Conferences. 90:01019.
Publisher | Google Scholor - Gibson, A.D., Yale, G., Corfmat, J. et al. (2022). Elimination of Human Rabies in Goa, India Through an Integrated One Health Approach. Nat Commun. 13:2788.
Publisher | Google Scholor - Naser RS, Lam MC, Qamar F, Zaidan BB. (2023). Smartphone-Based Indoor Localization Systems: A Systematic Literature Review. Electronics. 12(8):1814.
Publisher | Google Scholor - Mogano K, Suzuki T, Mohale D, Phahladira B, Ngoepe E, et al. (2022). Spatio-Temporal Epidemiology of Animal and Human Rabies in Northern South Africa Between 1998 And 2017. PLoS Negl Trop Dis. 16(7):e0010464.
Publisher | Google Scholor - N. Yau, (2012). A Guide to Choropleth Maps.
Publisher | Google Scholor - T. Przytycka, Lecture 11 Phylogenetic trees Phylogenetic (evolutionary) Tree.
Publisher | Google Scholor - Guo, D., Yin, W., Yu, H. et al. (2018). The Role of Socioeconomic and Climatic Factors in The Spatio-Temporal Variation of Human Rabies in China. BMC Infect Dis, 18:526.
Publisher | Google Scholor - Hall, Alastair. (2007). Generalized Method of Moments.
Publisher | Google Scholor - Dubé, Jean, Legros, Diègo. (2014). Spatial Autocorrelation.
Publisher | Google Scholor - Ismail MZ, AL- Hamdi NK, AL- Amery AN, Marston DA, McElhinney L, et al. (2020). Quantifying and Mapping the Burden of Human and Animal Rabies in Iraq. PLoS Negl Trop Dis. 14(10): e0008622.
Publisher | Google Scholor - Mounir Khayli, Youssef Lhor, Mohammed Bengoumi, Khalil Zro, Mehdi El Harrak, et al. (2021). Using Geostatistics to Better Understand the Epidemiology of Animal Rabies in Morocco: What Is the Contribution of The Predictive Value? Heliyon, 7(1):e06019.
Publisher | Google Scholor - Kleijnen, Jack. (2017). Kriging: Methods and Applications. SSRN Electronic Journal.
Publisher | Google Scholor - Vargas A, Romano APM, Merchán-Hamann E. (2019). Human Rabies in Brazil: A Descriptive Study, 2000-2017. Epidemiol Serv Saude. 28(2):e2018275.
Publisher | Google Scholor - R. Chattamvelli, R. Shanmugam, (2023). Descriptive Statistics, Synth. Lect. Math. Stat., 5:1-34.
Publisher | Google Scholor - Gebru G, et al. (2019). Risk Factors and Spatio-Temporal Patterns of Human Rabies Exposure in Northwestern Tigray, Ethiopia. Annals of Global Health. 85(1):1-12.
Publisher | Google Scholor - Babak, O., Deutsch, C.V. (2009). Statistical approach to inverse distance interpolation. Stoch Environ Res Risk Assess. 23:543-553.
Publisher | Google Scholor - Kadowaki H, Hampson K, Tojinbara K, Yamada A, Makita K. (2018). The risk of rabies spread in Japan: a mathematical modelling assessment. Epidemiology and Infection. 146(10):1245-1252.
Publisher | Google Scholor - Shoukri, M., Aleid, M. (2022). Quasi-Negative Binomial: Properties, Parametric Estimation, Regression Model and Application to RNA-SEQ Data. Open Journal of Statistics, 12:216-237.
Publisher | Google Scholor - Yamagata J, Ahmed K, Khawplod P, Mannen K, Xuyen DK, et al. (2007). Molecular Epidemiology of Rabies in Vietnam. Microbiol Immunol. 51(9):833-840.
Publisher | Google Scholor - M. G. Martynov, (1998). Spatial Access Methods, Program. Comput. Softw., 24(3):137-143.
Publisher | Google Scholor - Constantino, C.; Da Silva, E.C.; Dos Santos, D.M.; Paploski, I.A.D.; Lopes, M.O.; et al. (2023). One Health Approach on Dog Bites: Demographic and Associated Socioeconomic Factors in Southern Brazil. Trop. Med. Infect. Dis. 8:189.
Publisher | Google Scholor - Ribeiro, J., Staudacher, C., Martins, C.M. et al. (2018). Bat Rabies Surveillance and Risk Factors for Rabies Spillover in An Urban Area of Southern Brazil. BMC Vet Res. 14:173.
Publisher | Google Scholor - A. Horawa, L -functions, 1-79.
Publisher | Google Scholor - J. Burt, G. Barber, (1996). Spatial Statistic Standard Distance, Elem. Stat. Geogr.
Publisher | Google Scholor - A. Unwin, (1996). Geary’s Contiguity Ratio, Econ. Soc. Rev. (Irel)., 27(2):145-159.
Publisher | Google Scholor - Shim SR, Kim SJ, Lee J. (2019). Diagnostic Test Accuracy: Application and Practice Using R Software. Epidemiol Health. 41:e2019007.
Publisher | Google Scholor - Cazemier JL, Clascá F, Tiesinga PHE. (2016). Connectomic Analysis of Brain Networks: Novel Techniques and Future Directions. Front. Neuroanatomy. 10:110.
Publisher | Google Scholor - JABAROV Jahandar. (2023). Bias In Scientific Research: How to Identify and Eliminate It. Journal of Science and İnnovative Technologies. 25:80--96.
Publisher | Google Scholor - Amaral Mali M, Machado FN, Moniz FP, Bosco Alves Dos Santos F, Laot PAME, et al. (2024). The First Confirmed Human Case of Rabies, Timor-Leste, 2024. Euro Surveill. 29(18):2400241.
Publisher | Google Scholor - Rabies updates in Indonesia, Towar. Rabies Elimin. ASEAN, 9 (2018).
Publisher | Google Scholor - G. Cao, (2013). Lecture 6: Spatial Analysis and Modeling (GIST 4302/5302), Depts. Ttu. Edu, 66.
Publisher | Google Scholor - L. Anselin, (2017). The GeoDa Book Exploring: Spatial Data, 15.
Publisher | Google Scholor - Leigh, J., Girado, J., Singh, R., Johnson, A., Park, K., et al, (2002). TeraVision: a Platform and Software Independent Solution for Real Time Display Distribution in Advanced Collaborative Environments, Access Grid Retreat 2002 Proceedings, La Jolla, CA.
Publisher | Google Scholor - Introduction, “A, d, r”.
Publisher | Google Scholor - Acharya KP, Subedi D, Wilson RT. (2021). Rabies Control in South Asia Requires a One Health Approach. One Health. 12:100215.
Publisher | Google Scholor